淀粉是果实重要的贮藏碳水化合物,由直链淀粉(10%~25%)和支链淀粉(75%~90%)两类葡萄糖聚合物组成[1-2]。前者为由葡萄糖残基以α -1,4-糖苷键首尾聚合而成的无分支的直链分子,后者则是葡萄糖残基以α-1,4-糖苷键及α-1,6-糖苷键聚合而成的带分支长链。非晶体结构的直链淀粉和支链淀粉相互作用形成的晶体结构最终形成不溶于水的半晶体颗粒,即淀粉粒[3-4]。淀粉按功能可分为暂时淀粉和贮藏淀粉,暂时淀粉分布在叶片等“源”组织中,白天光合作用形成暂时淀粉,晚上暂时淀粉降解,为叶片呼吸作用和进一步的蔗糖合成提供底物并转移到植物的“库”[5-6]。
贮藏淀粉分布在果实、种子、块根、块茎、鳞茎等非光合作用细胞质体(造粉体)中,用于中期或长期贮藏,当植物需要时,会重新降解利用,如谷物类种子萌发所需的能量主要由贮藏的淀粉水解获得[7-9]。某些跃变型果实在成熟前含有大量的贮藏淀粉,如香蕉[10]、猕猴桃[11-12]、芒果[13]、鳄梨[14]等,在采后成熟过程中淀粉降解作为果实提供呼吸代谢的底物并形成风味影响果实的食用品质。
淀粉降解的主要产物为麦芽糖和葡萄糖,它们都可以被运送至质体外为细胞代谢利用[15]。淀粉的降解为非光合器官维持持续的碳水化合物供应,对于植物正常生长和适应生长环境至关重要[2]。果实淀粉降解对于果实风味品质形成,尤其是呼吸跃变型果实,在采后成熟过程中,淀粉含量会随着呼吸跃变急剧下降[10]。为此,我们近年来几种高贮藏淀粉的跃变型果实淀粉降解的研究进展,概括其淀粉降解参与的酶基因及其存在的转录调控因子,以期了解果实贮藏淀粉降解的特点,为今后果实淀粉降解基础研究提供理论依据和研究方向。
1 植物暂时淀粉降解
在模式植物拟南芥叶片淀粉降解研究表明,叶片暂时淀粉降解由多种酶蛋白共同参与完成(图1)[16],这些酶基因缺失或沉默都会导致淀粉过量表型,主要包括:葡聚糖水合双激酶(Glucan water dikinase,GWD)[17]、磷酸化葡聚糖水合双激酶(Phosphoglucan water dikinase,PWD)[18]、双特异性磷酸酶(Dualspecificity phosphate,DSP4,又名Starch excess 4,SEX4)[19]、磷酸化酶LSF1/2 (Like SEX Four 1/2)[20]、α-淀粉酶(α-amylase,AMY)[21]、β-淀粉酶(β-amylase,BAM)[22-23]、异淀粉酶(isoamylase,ISA)[24]、极限糊精酶(Limit dextrinase,LDA)、α-葡聚糖磷酸化酶(α-glucan phosphorylase,PHS)[25]、4-α葡聚糖转移歧化酶(4-α-glucanotransferase Disproportionating enzyme,DPE)[26-27]、麦芽糖转运蛋白(Maltose transporter,MEX)[28]及葡萄糖转运蛋白(Glucose transporter,pGlcT)[29]以及淀粉粒相关功能蛋白早期饥饿蛋白(early starvation1,ESV1)和类早期饥饿蛋白(Like Early Starvation1,LESV)[30]等。拟南芥叶片淀粉降解的基础研究为其他模式植物以及果实贮藏淀粉降解提供充足的参考价值。

图1 拟南芥叶片淀粉降解示意图
Fig.1 Sketch map of starch degradation in Arabidopsis leaves
2 贮藏淀粉降解研究
果实是植物贮藏淀粉的重要器官之一,如未成熟的香蕉、猕猴桃、苹果等跃变型果实中都含有大量淀粉[12, 31- 32]。淀粉在果实生长发育期慢慢积累并在成熟过程中降解为可溶性糖,使果实甜度上升。目前报道参与果实淀粉降解的酶基因只在番茄、猕猴桃、苹果和香蕉等水果中少数报道(表1),但相关的遗传规律和调控机制尚无深入研究。
表1 跃变型果实淀粉降解相关基因汇总
Table 1 Summary of genes related to starch degradation in climacteric fruits

与果实淀粉降解相关报道基因Genes related to starch degradation番茄 Tomato bAmy1/2/3[35]、SlGWD1[32]、SlPWD1[32]、SlLSF2[32]、SlBAM1[32]、SlAMY3[32]、SlAMY4[32]、苹果 Apple MdAM[39]猕猴桃 Kiwi fruit AdAMY1[42]、AdAGL3[42]、AdBAM3.1/3L/9[11,42]、AcPWD[43]、AcAMY1[43]、AcAMY3[43]、AcBAM1[43]、AcABAM3[43]、AdDof3[11]跃变型果实Climacteric fruit MAmy[47]、Maisa[48]、Ma-bmy[49]、MaGWD1[10]、MaPWD1[10]、MaSEX4[10]、MaLSF1[10]、MaLSF2[10]、MaBAM1/2/3/4/6/7/8/10[10,46,50]、MaAMY2B/2C/3/3A/ 3C[10,46,50]、MaISA2/3[10,46]、MaPHS2[10,50]、MaMEX1/2[10]、MapGlcT2-1/2-2/4-1/4-2[10]、MabHLH6 [10]、MaMYB3[32]其他 Other 芒果(comp44532、comp39992)[51],甜瓜(locus-12354、locus-15651)[52]、番木瓜(ko00500)[53]香蕉Banana
2.1 番茄果实淀粉降解
在番茄果实发育过程中,淀粉含量达到最大值后,淀粉开始水解,淀粉合成速度低于降解速度,当果实到达着色期(Color break)时,淀粉基本完全降解[33]。然而早期早番茄果实淀粉降解的相关研究未受到关注,在花粉淀粉降解的研究则有所进展,利用转座子插入技术突变表达番茄LeGWD基因(legwd:Ds),使得番茄雄性不育,突变后的番茄花粉显示出sex表型,抑制了花粉萌发,研究结果说明葡聚糖磷酸化酶参与花粉淀粉的降解过程[34]。后来果实淀粉降解的相关研究才开始被重视,MARIA等[35]发现β-淀粉酶活性随着番茄成熟淀粉降解及可溶性糖含量上升而提高,推测3个β-淀粉酶基因(bAmy1/2/3)参与淀粉的水解。最新研究发现,SlWHY1蛋白通过调节淀粉降解,增强番茄植株耐冷性但未见影响番茄果实淀粉降解报道[36]。而香蕉转录因子MaMYB3在Micro-Tom小番茄过表达结果显示,番茄果实中淀粉降解相关基因SlGWD1、SlPWD1、SlLSF2、SlBAM1,SlAMY3和 SlAMY4表达下调,淀粉降解受阻延缓果实成熟[32]。总而言之,番茄果实淀粉降解关键酶基因的挖掘及功能验证研究欠缺,还需进一步筛选验证。
2.2 苹果果实淀粉降解
研究发现,在苹果果实发育过程中,β-淀粉酶和α-淀粉酶活性活性变化与淀粉含量大致呈现互为消长的趋势,推测它们参与了果实质体中淀粉的降解过程[37-38]。而果实淀粉降解受到乙烯抑制剂1-MCP显著抑制,嘎拉苹果果实软化的初始阶段,淀粉降解对果实软化的影响最显著,并伴随淀粉酶(AM)活性和MdAM基因表达的快速上升,且淀粉含量、AM活性及它们与硬度的相关性均显著受到低温和乙烯因子的调节,表明淀粉降解与嘎拉苹果果实软化关系密切[39]。DOERFLINGER等[40]以两个苹果品种(McIntosh、Empire)为试材,在果实采收前1周处理1-MCP或2、4周前分别处理AVG,发现Empire果实采后贮藏过程中淀粉降解延迟,但不能阻止淀粉的最终降解。苹果作为我国北方主产水果之一,控制贮藏条件,果实可以进行将近一年时间的贮藏,贮藏过程中淀粉的降解与其耐贮性关系还需更多证据。
2.3 猕猴桃果实淀粉降解
猕猴桃果实采后成熟过程中淀粉降解,并伴随着可溶性糖(葡萄糖、果糖和蔗糖)含量上升[41-42]。采收后的果实淀粉含量在室温下随时间延续而下降,乙烯加剧淀粉的降解,而低温能有效延缓贮藏过程中淀粉水解酶活性上升从而有效缓解淀粉降解。研究发现,有24个基因与猕猴桃(Actinidia deliciosa)淀粉降解相关,其中AdAMY1,AdAGL3和AdBAM3.1/3L/9受乙烯诱导与淀粉降解正相关并受气调贮藏抑制,推测这5个基因参与猕猴桃采后成熟过程中淀粉的降解[42]。陈景丹等[43]研究发现相似结果,红阳猕猴桃(Actinidia chinensis)采后淀粉降解与果实软化密切相关,乙烯能通过调节AcPWD、AcAMY1、AcAMY3、AcBAM1和AcABAM3的表达,进而促进淀粉降解和果实软化,而外源1-MCP作用相反。最新研究发现在猕猴桃叶片中稳定过表达AdBAM3L能有效降低叶片淀粉含量,说明其是猕猴桃淀粉降解的关键酶基因,另外,转录因子AdDof3通过结合AdBAM3L的启动子并激活表达,从而参与果实淀粉降解的调控[11]。猕猴桃果实淀粉的降解与果内软化密切相关,不但在关键酶基因筛选上进行了探究,还在转录调控层面发现关键调控转录因子,丰富了果实淀粉降解的网络,为其他果实淀粉降解研究提供参考。
2.4 香蕉果实淀粉降解
香蕉果实发育过程中,淀粉慢慢积累起来,可达到干重的60%~90%[44-45],采后成熟过程中迅速降解转化为可溶性糖,淀粉含量从呼吸跃变前的22%鲜重降到跃变后的1%鲜重[46]。果肉淀粉降解是果实硬度下降的一个重要因素,因为淀粉不但作为细胞的主要贮藏物,还是呼吸能量源,为细胞呼吸跃变及乙烯诱发的高速细胞壁代谢提供前提条件,最终果实衰老软化。香蕉果肉淀粉降解的早期研究主要是发现与淀粉降解相关的酶基因:α-淀粉酶基因MAmy[47],淀粉去分支酶基因Maisa[48]和β-淀粉酶基因Mabmy[49]。近年研究表明β-淀粉酶在大蕉和巴西蕉果实淀粉降解中起关键作用[50]。另外最新转录水平和蛋白组研究发现,香蕉参与果肉淀粉降解的38酶基因,并有27个酶基因受乙烯诱导表达提高。同时发现MabHLH6能结合并激活 MaGWD1、MaLSF2、MaBAM1、MaBAM2、MaBAM8、MaBAM10、MaAMY3、MaAMY3C、MaISA2,MaISA3和MapGlcT2-2等11个淀粉降解相关基因的启动子,是响应果实成熟淀粉降解起关键作用的转录激活子[10]。后续研究进一步发现,一个MYB类转录因子MaMYB3可以靶定MabHLH6和10个淀粉降解相关酶基因的启动子,并抑制其转录表达。在番茄中超表达MaMYB3,发现番茄果实中淀粉降解相关基因表达下调,淀粉降解受阻延缓果实成熟,揭示了MaMYB3通过直接抑制淀粉降解相关基因和MabHLH6,参与调控果实成熟[32],丰富了香蕉降解的网络,如图2所示。
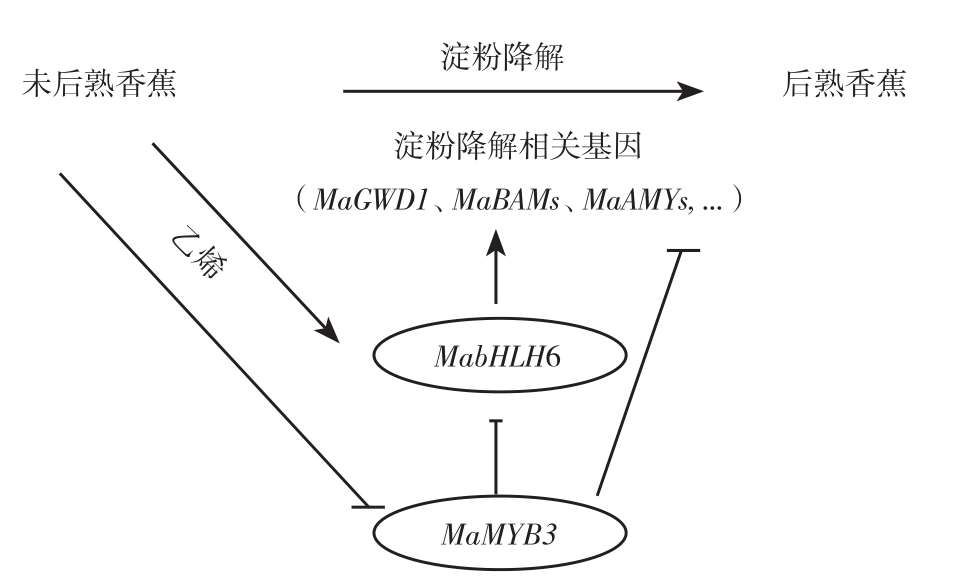
图2 香蕉淀粉降解调控网络
Fig.2 Degradation regulation network of banana starch
3 果实淀粉降解研究展望
在人们生活水平不断改善的今天,水果糖含量备受农业和消费者关注,因此关于各种果实在发育成熟和采后衰老过程中淀粉的降解调控作用机理是目前研究的热点。果实成长发育至采后成熟是一个动态的生命活动过程,前阶段果实主要是果实淀粉积累,后阶段以果实淀粉降解为主。果实淀粉降解由一系列酶基因参与完成,同时受果实成熟相关众多影响因素在转录水平和转录后水平调控。果实淀粉降解研究虽然起步较晚,但是依靠目前成熟的转录组测序、蛋白组测序、代谢组质谱分析和基因定点敲除等研究手段,未来十年,果实淀粉降解研究将迎来重大发展和突破。
3.1 果实淀粉降解研究的不足之处
作为果实研究的模式植物,番茄果实淀粉的降解研究存在较大空白,涉及果实淀粉降解的酶基因还未被完全挖掘。香蕉和猕猴桃作为高淀粉水果,淀粉降解的研究在近几年才有所突破,不过目前还未涉及遗传规律研究。果实淀粉降解的调控是一个复杂过程,涉及多种转录水平,转录后水平以及翻译后水平的多层次、多阶段的协同作用(图3)。因此目前研究某个或少数几个转录因子调控淀粉降解仍不足以阐明果实淀粉降解的复杂调控网络。

图3 多层次果实常数降解研究思路
Fig.3 Research idea of mutti-level degradation of fruit starch
3.2 果实淀粉降解与非果实淀粉降解的异同
非果实淀粉主要包括叶片暂时淀粉和组织贮藏淀粉两种存在形式。叶片暂时淀粉是叶片通过光合作用在叶绿体中合成临时储备碳源,并可通过降解为葡萄糖和麦芽糖长距离运输到植物其他部位。而组织贮藏淀粉多贮藏于块茎或块根细胞淀粉体和种子胚乳中,作为组织器官的营养物质,以备生长所需,如块茎和种子萌芽。如马铃薯淀粉体中的淀粉降解主要发生在以下两种情况:一种是马铃薯低温贮藏过程中淀粉降解糖化,导致还原糖积累。另外是贮藏淀粉降解为块茎发芽提供必需的营养物质[54]。淀粉降解主要通过淀粉磷酸化途径和淀粉水解,其过程包括可溶性葡聚糖的释放、可溶性和线性葡聚糖的降解和麦芽糖代谢3个阶段。目前研究发现马铃薯α-淀粉酶活性主要由StAmy23引起,β-淀粉酶活性主要由StBAM1和tBAM7引起,冷藏块茎中的淀粉酶抑制剂可以调节淀粉酶的活性[55]。
果实淀粉降解与叶片暂时淀粉降解具有更多相似之处,淀粉降解相关的酶在淀粉贮藏细胞中合成,参与淀粉降解相关的酶种类相似,但是叶片淀粉降解受光照调控启动,而果实淀粉降解不受光照影响,而受一系列激素(尤其是乙烯)诱导调控。另外,谷物种子胚乳是死细胞组织,降解淀粉所需的酶由盾片或者糊粉层(包围在胚乳表面的一层活细胞)合成并释放到胚乳中,又或者是β-淀粉酶在种子发育时就形成贮藏在胚乳中,后经种子萌发时被活化起作用。然而目前研究发现,只有三种淀粉降解相关的酶参与胚乳淀粉的降解,分别是:α-淀粉酶[56]、β-淀粉酶[57]、DBE(极限糊精酶,LD)[58]。虽然 GWD 和PWD都存在于谷类种子中,种子贮藏淀粉粒上只检测到非常低水平的共价连接的磷酸盐,也没有足够的ATP去维持循环的磷酸化作用[59]。因此,谷类淀粉降解与叶绿体暂时淀粉和果实贮藏淀粉的降解的机理有明显不同。
3.3 果实淀粉降解的调控研究思路
转录因子参与果实发育成熟的研究越来越受到关注,此类核小蛋白在果实成熟发育过程中起到杠杆效果[60]。在线转录因子数据库(PlantTFDB 4.0[61]和iTAK[62])并结合表达谱数据分析结果,可以让研究者获得大量的与果实降解同步表达差异转录因子,为后期阐明果实淀粉降解的复杂转录调控网络提供最基础的数据。
随着蛋白组测序技术的高速发展,翻译后水平的蛋白修饰调控也备受研究者青睐,如蛋白泛素化测序、组蛋白乙酰化测序、蛋白磷酸化测序等。蛋白质泛素化是体内具有重要生物学功能的蛋白质翻译后修饰之一,体内泛素化和去泛素化的动态平衡,是调控蛋白质降解和维持细胞蛋白稳态的关键机制,对蛋白质的定位、代谢、功能调节和降解中都起着十分重要的作用[63]。蛋白质磷酸化是一种非常重要且广泛存在的翻译后修饰调控方式,参与细胞的增殖、发育、分化、凋亡,细胞骨架调控、新陈代谢等,对许多生物的细胞功能起着生物“开/关”作用[64]。蛋白质乙酰化参与了几乎所有的生物学过程,如转录、应激反应、新陈代谢以及蛋白合成与降解等,除了对核内转录调控因子的激活外,对蛋白质的功能也产生很大影响,包括酶的活化与失活、蛋白质稳定性、亚细胞结构定位和特殊功能复合体的形成等[65]。
同时,通过质谱分析,不但可以知道蛋白是否发生修饰,而且可以确定修饰的位点及程度。为果实淀粉降解参与调控的转录因子或蛋白的深入研究提供平台支持。
另外,定点突变技术CRISPR自2014在番茄中便开启应用,此后,CRISPR-Cas9系统已成功应用于柑橘、黄瓜、苹果、葡萄、西瓜、猕猴桃、香蕉等作物[66]。此技术的推广将为果实淀粉降解的研究提供强大的科研工具,将大大促进构建完善的果实淀粉降解的调控网络,挖掘关键的转录因子或者蛋白修饰调控组分。总而言之,研究果实淀粉降解,将丰富果实采前和采后生物学的理论,有助于利用基因工程或者其他手段改进果实的保鲜技术。
[1]YANG S T,LIU X J,QIAO S,TAN W F,LI M,FENG J Y,ZHANG C,KANG X,HUANG T B,ZHU Y L,YANG L,WANG D.Starch content differences between two sweet potato accessions are associated with specific changes in gene expression[J].Functional & Integrative Genomics,2018,18(6):613-625.doi:10.1007/s10142-018-0611-2.
[2]刘襄河,郑丽璇,郑丽勉,欧成成,叶超霞,王安利.双波长法测定常用淀粉原料中直链淀粉、支链淀粉及总淀粉含量[J].广东农业科学,2013,40(18):97-100.doi:10.3969/j.issn.1004-874X.2013.18.034.
LIU X H,ZHENG L X,ZHENG L M,OU C C,YE C X,WANG A L.Determination of amylose and amylopectin in the commonly used starch materials by dual-wavelength spectrophotometry[J].Guangdong Agricultural Sciences,2013,40(18):97-100.doi:10.3969/j.issn.1004-874X.2013.18.034.
[3]MATSUSHIMA R,MAEKAWA M,KUSANO M,TOMITA K,KONDO H,NISHIMURA H,CROFTS N,FUJITA N,SAKAMOTO W.Amyloplast membrane protein SUBSTANDARD STARCH GRAIN6 controls starch grain size in rice endosperm[J].Plant Physiology,2016,170(3):1445-1459.doi:10.1104/pp.15.01811.
[4]SEUNG D,SMITH A M.Starch granule initiation and morphogenesisprogress in Arabidopsis and cereals[J].Journal of Experimental Botany,2019,70(3):771-784.doi:10.1093/jxb/ery412.
[5]MALINOVA I,QASIM H M,BRUST H,FETTKE J.Parameters of starch granule genesis in chloroplasts of Arabidopsis thaliana[J].Frontiers in Plant Science,2018,9:761.doi:10.3389/fpls.2018.00761.
[6]KÖTTING O,KOSSMANN J,ZEEMAN S C,LLOYD J R.Regulation of starch metabolism: the age of enlightenment?[J].Current Opinion in Plant Biology,2010,13(3):321-329.doi:10.1016/j.pbi.2010.01.003.
[7]何秀英,伍时照,宋美芳,林贤琛.水稻籽粒发育和胚乳淀粉粒形成的研究[J].广东农业科学,2000(2):8-10.doi:10.3969/j.issn.1004-874X.2000.02.004.
HE X Y,WU S Z,SONG M F,LING X S.Study on development of seed and formation of starch in endosperm of rice[J].Guangdong Agricultural Sciences,2000(2):8-10.doi:10.3969/j.issn.1004-874X.2000.02.004.
[8]DAMARIS R N,LIN Z,YANG P,HE D.The rice Alpha-Amylase,conserved regulator of seed maturation and germination[J].International Journal of Molecular Sciences,2019,20(2).doi:10.3390/ijms20020450.
[9]DING J,HOU G G,NEMZER B V,XIONG S,DUBAT A,FENG H.Effects of controlled germination on selected physicochemical and functional properties of whole-wheat flour and enhanced gammaaminobutyric acid accumulation by ultrasonication[J].Food Chemistry,2018,243:214-221.doi:10.1016/j.foodchem.2017.09.128.
[10]XIAO Y Y,KUANG J F,QI X N,YE Y J,WU Z X,CHEN J Y,LU W J.A comprehensive investigation of starch degradation process and identification of a transcriptional activator MabHLH6 during banana fruit ripening[J].Plant Biotechnology Journal,2018,16(1):151-164.doi:10.1111/pbi.12756.
[11]ZHANG A D,WANG W Q,TONG Y,LI M J,GRIERSON D,FERGUSON I,CHEN K S,YIN X R.Transcriptome analysis identifies a zinc finger protein regulating starch degradation in Kiwifruit[J].Plant Physiology,2018,178(2):850-863.doi:10.1111/nph.15532.
[12]LI D,ZHU F.Starch structure in developing kiwifruit[J].International Journal of Biological Macromolecules,2018,120(A):1306-1314.doi:10.1016/j.ijbiomac.2018.08.128.
[13]CALDERON C A,VEGA G M O,DE J,ZAZUETA M J,FITCH V P R,CARRILLO L A,GUTIERREZ D R,LIMON V V,AGUILAR P E.Effect of extrusion process on the functional properties of high amylose corn starch edible films and its application in mango (Mangifera indica L.) cv.Tommy Atkins[J].Journal of Food Science and Technologymysore,2018,55(3):905-914.doi:10.1007/s13197-017-2997-6.
[14]ALCARAZ M L,HORMAZA J I,RODRIGO J.Ovary starch reserves and pistil development in avocado (Persea americana)[J].Physiology Plant,2010,140(4):395-404.doi:10.1111/j.1399-3054.2010.01410.x.
[15]MCCOY J G,REN Z N,STANEVICH V,LEE J,MITRA S,LEVIN E J,POGET S,QUICK M,IM W,ZHOU M.The Structure of a sugar transporter of the glucose EIIC superfamily provides insight into the elevator mechanism of membrane transport[J].Structure,2016,24(6):956-964.doi:10.1016/j.str.2016.04.003.
[16]SILVER D M,KOTTING O,MOORHEAD G B.Phosphoglucan phosphatase function sheds light on starch degradation[J].Trends in Plant Science,2014,19(7):471-478.doi:10.1016/j.tplants.2014.01.008.
[17]SKEFFINGTON A W,GRAF A,DUXBURY Z,GRUISSEM W,SMITH A M.Glucan,water dikinase exerts little control over starch degradation in Arabidopsis Leaves at Night[J].Plant Physiology,2014,165(2):866-879.doi:10.1104/pp.114.237016.
[18]KÖTTING O,PUSCH K,TIESSEN A,GEIGENBERGER P,STEUP M,RITTE G.Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves[J].Plant Physiology,2005,137(1):242-252.doi:10.1104/pp.104.055954.
[19]OLIVER K T,DIANA S,CHRISTOPH E,SIMONA E,TINA M,GENTRY M S,SYLVIANE C M,JYCHIAN C,SMITH A M,MARTIN S.Starch-Excess4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana[J].The Plant Cell,2009,21(1):334-346.doi:10.1105/tpc.108.064360.
[20]WILKENS C,AUGER K D,ANDERSON N T,MEEKINS D A,RATHTHAGALA M,ABOU H M,PAYNE C M,GENTRY M S,SVENSSON B.Plant alpha-glucan phosphatases SEX4 and LSF2 display different affinity for amylopectin and amylose[J].Febs Letters,2016,590(1):118-128.doi:10.1002/1873-3468.12027.
[21]YU T S,ZEEMAN S C,THORNEYCROFT D,FULTON D C,DUNSTAN H,LUE W L,HEGEMANN B,TUNG S Y,UMEMOTO T,CHAPPLE A,TSAI D L,WANG S M,SMITH A M,CHEN J,SMITH S M.Alpha-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves[J].Journal of Biological Chemistry,2005,280(11):9773-9779.doi:10.1074/jbc.M413638200.
[22]MONROE J D,STORM A R.Review: The Arabidopsis beta-amylase(BAM) gene family: Diversity of form and function[J].Plant Science,2018,276:163-170.doi:10.1016/j.plantsci.2018.08.016.
[23]LI J,ZHOU W,FRANCISCO P,WONG R,ZHANG D,SMITH S.M.Inhibition of Arabidopsis chloroplast beta-amylase BAM3 by maltotriose suggests a mechanism for the control of transitory leaf starch mobilisation[J].PLoS One,2017,12(2):e172504.doi:10.1371/journal.pone.0172504.
[24]SUNDBERG M,PFISTER B,FULTON D,BISCHOF S,DELATTE T,EICKE S,STETTLER M,SMITH S M,STREB S,ZEEMAN S C.The heteromultimeric debranching enzyme involved in starch synthesis in Arabidopsis requires both isoamylase1 and isoamylase2 subunits for complex stability and activity[J].PLoS One,2013,8(9):e75223.doi:10.1371/journal.pone.0075223.
[25]SCHWARTE S,WEGNER F,HAVENSTEIN K,GROTH D,STEUP M,TIEDEMANN R.Sequence variation,differential expression,and divergent evolution in starch-related genes among accessions of Arabidopsis thaliana[J].Plant Molecular Biology,2015,87(4-5):489-519.doi:10.1007/s11103-015-0293-2.
[26]MALINOVA I,FETTKE J.Reduced starch granule number per chloroplast in the dpe2/phs1 mutant is dependent on initiation of starch degradation[J].PLoS One,2017,12(11):e187985.doi:10.1371/journal.pone.0187985.
[27]SMIRNOVA J,FERNIE A R,SPAHN C M T,STEUP M.Photometric assay of maltose and maltose-forming enzyme activity by using 4-alpha-glucanotransferase (DPE2) from higher plants[J].Analytical Biochemistry,2017,532:72-82.doi:10.1016/j.ab.2017.05.026.
[28]REIDEL E J,TURGEON R,CHENG L.A maltose transporter from apple is expressed in source and sink tissues and complements the Arabidopsis maltose export-defective mutant[J].Plant and Cell Physiology,2008,49(10):1607-1613.doi:10.1093/pcp/pcn134.
[29]CHO M H,LIM H,SHIN D H,JEON J S,BHOO S H,PARK Y I,HAHN T R.Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana[J].New Phytologist,2011,190(1):101-112.doi:10.1111/j.1469-8137.2010.03580.x.
[30]FEIKE D,SEUNG D,GRAF A,BISCHOF S,ELLICK T,COIRO M,SOYK S,EICKE S,METTLER A T,LU K J,TRICK M,ZEEMAN S C,SMITH A M.The starch granule-associated protein EARLY STARVATION1 is required for the control of starch degradation in Arabidopsis thaliana leaves[J].The Plant Cell,2016,28(6):1472-1489.doi:10.1105/tpc.16.00011.
[31]LI D,ZHU F.Physicochemical properties of kiwifruit starch[J].Food Chemistry,2017,220:129-136.doi:10.1016/j.foodchem.2016.09.192.
[32]FAN Z Q,BA L J,SHAN W,XIAO Y Y,LU W J,KUANG J F,CHEN J Y.A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6[J].The Plant Journal,2018,96(6):1191-1205.doi:10.1111/tpj.14099.
[33]LUENGWILAI K,BECKLES D M.Structural investigations and morphology of tomato fruit starch[J].Journal of Agricultural and Food Chemistry,2009,57(1):282-291.doi:10.1021/jf802064w.
[34]NASHILEVITZ S,MELAMED B C,AHARONI A,KOSSMANN J,WOLF S,LEVY A A.The legwd mutant uncovers the role of starch phosphorylation in pollen development and germination in tomato[J].The Plant Journal,2009,57(1):1-13.doi:10.1111/j.1365-313X.2008.03664.x.
[35]MARIA T,TSANIKLIDIS G,DELIS C,NIKOLOPOULOU A E,NIKOLOUDAKIS N,KARAPANOS I,AIVALAKIS G.Gene transcript accumulation and enzyme activity of β-amylases suggest involvement in the starch depletion during the ripening of cherry tomatoes[J].Plant Gene,2016,5:8-12.doi:10.1016/j.plgene.2015.10.004.
[36]ZHUANG K,KONG F,ZHANG S,MENG C,YANG M,LIU Z,WANG Y,MA N,MENG Q.Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation[J].New Phytologist,2019,221(4):1998-2012.doi:10.1111/nph.15532.
[37]王永章,张大鹏.发育过程中苹果果实的β-淀粉酶:活性、数量变化和亚细胞定位[J].中国科学C辑,2002,32(3):201-210.doi:10.3321/j.issn:1006-9259.2002.03.002.
WANG Y J,ZHANG D P.Activity,quantitative changes,and subcellular localization of beta amylase in apple fruits during development[J].Science in China series (C),2002,32(3):201-210.doi:10.3321/j.issn:1006-9259.2002.03.002.
[38]王永章,张大鹏.苹果果实发育过程中α-淀粉酶的活性、数量变化和亚细胞定位[J].植物学报,2002,44(1):34-41.doi:10.3321/j.issn:1672-9072.2002.01.006.
WANG Y J,ZHANG D P.Activity,quantitative changes,and subcellular localization of alpha-amylase during development of apple fruits [J].Acta Botanica Sinica,2002,44(1):34-41.doi:10.3321/j.issn:1672-9072.2002.01.006.
[39]魏建梅,齐秀东,闫芳教.采后‘嘎拉’苹果果实糖和淀粉代谢及关键酶基因表达特性[J].北方园艺,2015(19):126-131.doi:10.11937/bfyy.201519031.
WEI J M,QI X D,YAN F J.Characteristics of sugar and starch metabolism and key enzyme gene expression in postharvest 'gala' apple fruit[J].Northern Horticulture,2015(19):126-131.doi:10.11937/bfyy.201519031.
[40]DOERFLINGER F C,NOCK J F,MILLER W B,WATKINS C B.Preharvest aminoethoxyvinylglycine (AVG) and 1-methylcyclopropene (1-MCP) effects on ethylene and starch concentrations of‘Empire’and‘McIntosh’apples[J].Scientia Horticulturae,2019,244:134-140.doi:https://doi.org/10.1016/j.scienta.2018.09.031.
[41]WEGRZYN,MACRAE.Alpha-amylase and starch degradation in kiwifruit[J].Journal of Plant Physiology,1995,147(1):19-28.doi:10.1016/S0176-1617(11)81407-0.
[42]HU X,KUANG S,ZHANG A D,ZHANG W S,CHEN M J,YIN X R,CHEN K S.Characterization of starch degradation related genes in postharvest Kiwifruit[J].International Journal of Molecular Sciences,2016,17(12).doi:10.3390/ijms17122112.
[43]陈景丹,许凤,陈伟,杨震峰.猕猴桃果实采后软化期间淀粉降解关键基因表达分析[J].核农学报,2018,32(2):236-243.doi:10.11869/j.issn.100-8551.2018.02.0236.
CHEN J D,XU F,CHEN W,YANG Z F.Expression analysis of key genes for starch degradation in kiwi fruit during postharvest softening[J].Journal of Nuclear Agricultural Sciences,2018,32(2):236-243.doi:10.11869/j.issn.100-8551.2018.02.0236.
[44]苗红霞,孙佩光,刘菊华,金志强,徐碧玉.香蕉果实可溶性淀粉合成酶基因MaSSⅢ-1的分子特征与时空表达模式[J].中国农学通报,2019,35(1):75-79.
MIAO H X,SUN P G,LIU J H,JING Z Q,XU B Y.The soluble starch synthase gene of banana fruit MaSS Ⅲ-1 molecular characteristics and expression pattern of time and space[J].Chinese Agricultural Science Bulletin,2019,35(1):75-79.
[45]陈海强,杨公明,梅为云,李昌宝,余铭.不同品种香蕉果实成熟期主要营养与功能成分含量变化[J].广东农业科学,2014,41(22):24-28.doi:10.3969/j.issn.1004-874X.2014.22.006.
CHEN H Q,YANG G M,MEI W H,LI C B,YU M.Changes of main nutritional and functional components in different varieties of banana during ripening[J].Guangdong Agricultural Sciences,2014,41(22):24-28.doi:10.3969/j.issn.1004-874X.2014.22.006.
[46]JOURDA C,CARDI C,GIBERT O,GIRALDO T A,RICCI J,MBEGUIE A D,YAHIAOUI N.Lineage-specific evolutionary histories and regulation of major starch metabolism genes during banana ripening[J].Frontiers in Plant Science,2016,7:1778.doi:10.3389/fpls.2016.01778.
[47]JUNIOR A V,DO N J R,LAJOLO F M.Molecular cloning and characterization of an alpha-amylase occurring in the pulp of ripening bananas and its expression in Pichia pastoris[J].Journal of Agricultural and Food Chemistry,2006,54(21):8222-8228.doi:10.1021/jf060805b.
[48]BIERHALS J D,LAJOLO F M,CORDENUNSI B R,DO N J.Activity,cloning,and expression of an isoamylase-type starch-debranching enzyme from banana fruit[J].Journal of Agricultural and Food Chemistry,2004,52(24):7412-7418.doi:10.1021/jf049300g.
[49]DO N J,JUNIOR A V,BASSINELLO P Z,CORDENUNSI B R,MAINARDI J A,PURGATTO E,LAJOLO F M.Beta-amylase expression and starch degradation during banana ripening[J].Postharvest Biology and Technology,2006,40(1):41-47.doi:10.1016/j.postharvbio.2005.11.008.
[50]GAO H J,HUANG S B,DONG T ,YANG Q S,YI G J.Analysis of resistant starch degradation in postharvest ripening of two banana cultivars: Focus on starch structure and amylases[J].Postharvest Biology and Technology,2016,119:1-8.doi:10.1016/j.postharvbio.2016.03.022.
[51]DAUTT C M,OCHOA L A,CONTRERAS V C.A,PACHECO S M A,CASAS F S,SANCHEZ F A,KUHN D N,ISLAS O M A.Mango(Mangifera indica L.) cv.Kent fruit mesocarp de novo transcriptome assembly identifies gene families important for ripening[J].Frontiers in Plant Science,2015,6:62.doi:10.3389/fpls.2015.00062.
[52]SHIN A Y,KIM Y M,KOO N,LEE S M,NAHM S,KWON S Y.Transcriptome analysis of the oriental melon (Cucumis melo L.var.makuwa) during fruit development[J].Peer J,2017,5:e2834.doi:10.7717/peerj.2834.
[53]ZHU X Y,YE L L,DING X C,GAO Q Y,XIAO S L,TAN Q Q,HUANG J L,CHEN W X,LI X P.Transcriptomic analysis reveals key factors in fruit ripening and rubbery texture caused by 1-MCP in papaya[J].BMC Plant Biology,2019,19(1):309.doi:10.1186/s12870-019-1904-x.
[54]何虎翼,唐洲萍,杨鑫,樊吴静,谭冠宁,李丽淑,何新民.马铃薯淀粉合成与降解研究进展[J].生物技术通报,2019,35(4):101-107.doi:10.13560/j.cnki.biotech.bull.1985.2018-0829.
HE H Y,TANG Z P,YANG X,FAN W J,TAN G N,LI L S,HE X M.Research progress on synthesis and degradation of potato starch[J].Biotechnology Bulletin,2019,35(4):101-107.doi:10.13560/j.cnki.biotech.bull.1985.2018-0829.
[55]ZHANG H L,HOU J,LIU J,XIE C H,SONG B T.Amylase analysis in potato starch degradation during cold storage and sprouting[J].Potato Research,2014,57(1):47-58.doi:10.1007/s11540-014-9252-6.
[56]BAK J K S,LAUGESEN S,OSTERGAARD O,FINNIE C,ROEPSTORFF P,SVENSSON B.Spatio-temporal profiling and degradation of alpha-amylase isozymes during barley seed germination[J].FEBS Journal,2007,274(10):2552-2565.doi:10.1111/j.1742-4658.2007.05790.x.
[57]URAIPONG C,ZHAO J.Rice bran protein hydrolysates exhibit strong in vitro alpha-amylase,beta-glucosidase and ACE-inhibition activities[J].Journal of the Science of Food and Agriculture,2016,96(4):1101-1110.doi:10.1002/jsfa.7182.
[58]MOLLER M S,WINDAHL M S,SIM L,BOJSTRUP M,ABOU H M,HINDSGAUL O,PALCIC M,SVENSSON B,HENRIKSEN A.Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase[J].Journal of Molecular Biology,2015,427(6 Pt B):1263-1277.doi:10.1016/j.jmb.2014.12.019.
[59]BLENNOW A,ENGELSEN S B,MUNCK L,MOLLER B L.Starch molecular structure and phosphorylation investigated by a combined chromatographic and chemometric approach[J].Carbohydrate Polymers,2000,41(2):163-174.doi:10.1016/S0144-8617(99)00082-X.
[60]范中奇,邝健飞,陆旺金,陈建业.转录因子调控果实成熟和衰老机制研究进展[J].园艺学报,2015,42(9):1649-1663.doi:10.16420/j.issn.0513-353x.2015-0356.
FAN Z Q,KUANG J F,LU W J,CHEN J Y.Advances in research of the mechanism of transcription factors involving in regulating fruit ripening and senescence[J].Acta Horticulturae Sinica,2015,42(9):1649-1663.doi:10.16420/j.issn.0513-353x.2015-0356.
[61]JIN J P,TIAN F,YANG D C,MENG Y Q,KONG L,LUO J C ,GAO G.PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants[J].Nucleic Acids Research,2017,45(1):1040-1045.doi:10.1093/nar/gkw982.
[62]ZHENG Y,JIAO C,SUN H,ROSLI H G,POMBO M A,ZHANG P,BANF M,DAI X,MARTIN G B,GIOVANNONI J J,ZHAO P X,RHEE S Y,FEI Z.ITAK: a program for genome-wide prediction and classification of plant transcription factors,transcriptional regulators,and protein kinases[J].Molecular Plant,2016,9:1667-1670.doi:10.1016/j.molp.2016.09.014.
[63]KELLEY D.E3 ubiquitin ligases: key regulators of hormone signaling in plants[J].Molecular & Cellular Proteomics,2018,17(6):117-476.doi:10.1074/mcp.MR117.000476.
[64]WANG P C,ZHAO Y ,LI Z P,HSU C C,LIU X,FU L W,HOU Y J,DU Y Y,XIE S J,ZHANG C G.Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response[J].Molecular Cell,2018,69(1):100-112.doi:10.1016/j.molcel.2017.12.002.
[65]WALLEY J W,SHEN Z X,MCREYNOLDS M R,SCHMELZ E A,BRIGGS S P.Fungal-induced protein hyperacetylation in maize identified by acetylome profiling[J].Proceedings of the National Academy of Sciences of the United States of America,2017,115(1):201717519.doi:10.1073/pnas.1717519115.
[66]WANG T,ZHANG H Y,ZHU H L.CRISPR technology is revolutionizing the improvement of tomato and other fruit crops[J].Horticulture Research,2019,6(1):77.doi:10.1038/s41438-019-0159-x.