【Research significance】Sugarcane streak mosaic virus(SCSMV), a member of the genus Poacevirus in the Potyviridae family[1-2], was first reported by Hall et al.in 1998[3].Diseases caused by this virus have caused significant sugarcane yield losses in many countries, including India,Bangladesh, Indonesia, Sri-Lanka, USA, Australia,Vietnam, Thailand and China[2, 4-13].SCSMV is one of the major pathogens causing mosaic disease in sugarcane[other two viruses are Sugarcane mosaic virus(SCMV)and Sorghum mosaic virus(SrMV)].It can be transmitted by plant extracts and sugarcane setts, while insect vectors clearly responsible for its transmission have not yet been identified[1,10].The virus now is a serious threat to sugarcane production in many sugarcane-producing regions.Therefore, there is an urgent need to establish an accurate and efficient detection method for the forecasting and controlling of SCSMV.【Research progress】A number of diagnostic methods have been developed for the detection of SCSMV, including serological test, double antibody sandwich enzyme-linked immunosorbent assay(DAS-ELISA)and direct antigen coating(DAC)-ELISA; reverse transcription-polymerase chain reaction(RT-PCR); immunocapture-RTPCR(IC-RT-PCR)and Duplex-Immunocapture(Duplex-IC)RT-PCR, one-step quadruplex RTPCR, colloidal gold immunochromatographic strip assay, reverse transcription recombinase polymerase amplification(RT-RPA)and quantitative realtime PCR[4,6,14-20].These methods, one way or the other, have their own intrinsic disadvantages, which include being time-consuming, with low sensitivity,insufficient specificity and/or requirements of costly sophisticated instruments.【Research breakthrough point】 The loop-mediated isothermal amplification(LAMP)method was developed as a rapid nucleic acid detection method, which can be applied to the detection of genomes by using reverse transcriptase[21].Further, the LAMP and reverse transcription loop-mediated isothermal amplification(RT-LAMP)assays have been proven to be simple, sensitive, specific and inexpensive for the detection of different DNA and RNA viruses.Various viruses including those from human, animals and plants can be easily detected by using RT-LAMP[22-24].【Key problems to be solve】The adaptation of this method is introduced for the detection of SCSMV from total RNA isolated from sugarcane leaves by using a primer set based on the coat protein gene sequences.
1 Materials and methods
1.1 Source of viruses
Sugarcane leaves with mosaic symptoms and healthy leaves were collected from Nanning City and stored in -80℃ for use.
1.2 Total RNA extraction
Total RNA was extracted from sugarcane leaves which showed mosaic symptoms by using the TRIzol reagent according to the manufacturer’s instructions(Tiangen, CA, China).RNA samples were resuspended in 50 μL DEPC-treated water and the concentrations were measured by using a micro-spectrophotometer(Nanodrop 2000, Thermo Scientific, USA).
1.3 Primer designed for RT-LAMP
The sequences of several SCSMV genomes were downloaded from the GenBank and were aligned by the software vector NTI.The conserved coding regions of the SCSMV coat protein were selected(JX467699 and JN163911), and primers used for RT-LAMP were designed by using the Primer Explorer V4 software(http://primerexplorer.jp/elamp4.0.0/index.html)and default settings(Table 1).
Table 1 RT-LAMP primers designed for detection of SCSMV
Note: SCSMV sequences(Accession No.: JX467699 and JN163911)were used as reference sequences.Coat protein gene sequences were used for designing the RT-LAMP primers.
Primer name Length(bp)Genome position Sequence(5’-3’)SCSMV-F3 20 9125-9144 AAGTGGACGAAAGCTGTGTT SCSMV-B3 18 9298-9315 TTCCCGCTACGAACCGAG SCSMV-FIP 41 9198-9219 GGCCCCATTTCGAACTCCACTT 9151-9169 GGATGGAGCTGATGGGACA SCSMV-BIP 41 9230-9251 AACGCCAAACCTGGTATTCGCG 9277-9295 CTGAACCCACTTGTACGCC
1.4 Virus detection by RT-PCR
The RT-PCR reactions were performed by using the PrimeScript One Step RT-PCR Kit Ver.2(TAKARA).Primer sets B3/F3, SCMV-F/R and SrMV-F/R were used for the detection of SCSMV,SCMV and SrMV, respectively[25-26].Separate positive samples were used for RT-LAMP.The RTPCR reaction(2 μL of PrimeScript 1 Step Enzyme Mix, 12.5 μL of 2×1 Step Buffer, 2 μL of Template RNA, 1 μL each of B3/F3, the reaction was brought to 25 μL with RNase-free water)was carried out in a thermocycler(TP 350, TaKaRa, CA, China)with the following procedure: initial incubation(50 ℃ for 30 min, then 94 ℃ for 2 min), 35 cycles of 94 ℃ for 30 s, 60 ℃ for 30 s and 72 ℃ for 30 s, followed by a final extension at 72 ℃ for 10 min.RT-PCR products were separated by electrophoresis in 1.5% agarose gels(AMRESCO Inc., Solon, OH, USA)in 1×TAE buffer and detected by the fluorescent GelRedTM Nucleic Acid Gel Staining(Biotium, Inc., Hayward,CA, USA).
1.5 RT-LAMP assay
RT-LAMP was carried out by using total RNA from potentially infected samples.The concentration of total RNA was 167.9 ng/µL.The amplification was carried out in a 25 µL reaction containing 2µL of RNA, 0.2 µmol/L of F3, 0.2 µmol/L of B3,0.8 µmol/L of FIP, 0.8 µmol/L of BIP, 6 mmol/L of MgSO4, 0.8 mol/L of betaine(Sigma–Aldrich,St.Louis, MO, USA), 10 mmol/L of dNTPs, 1 µL of Bst DNA polymerase(8 U/µL, New England Biolabs, MA, USA), 0.5 µL of M-MLV reverse transcriptase(200 U/µL, Promega, USA), 0.5 µL of Recombinant RNase Inhibitor(40 U/µL, Takara,China), 20 mmol/L of Tris–HCl(pH 8.8),10 mmol/L of KCl, 10 mmol/L of(NH4)2SO4,0.1% Triton X-100, and DEPC-treated water.The mixture was incubated at 60 ℃ , 61 ℃ , 62 ℃ , 63℃ , 64 ℃ and 65 ℃ for 60 min followed by 5 min at 80 ℃.The RT-LAMP amplification products(3µL)were analyzed by agarose gel electrophoresis(1.5% agarose, TAE), which was followed by the fluorescent GelRedTM Nucleic Acid Gel Staining and the direct visual detection of the reaction tube after the addition of SYBR Green I(Solarbio, China;1∶1000 TE, V/V).
1.6 Specificity of RT-LAMP
To determine the specificity of RT-LAMP, the total RNA was extracted from healthy sugarcane leaves and leaves infected by SCSMV(detected by RTPCR), SCMV(detected by RT-PCR)or SrMV(confirmed by RT-PCR)was used to determine the specificity of the RT-LAMP method.Again, in each case, the products were separated by agarose gel electrophoresis(1.5% agarose, TAE)and visualized after staining.
1.7 Sensitivity comparison between RT-LAMP and RT-PCR
RT-LAMP and RT-PCR were carried out using various concentrations of total RNA as the template.Total RNA quantified by NanoDrop 1 000(Thermo Scientific, USA)was serially diluted 10-fold from 167.9 ng/µL to 167.9×10-8 ng/µL prior to being used as the template for RT-LAMP and RT-PCR.After the reaction was completed, the reaction products were resolved on 1.5% agarose gels and visualized after staining.
1.8 Detection of field samples by RT-LAMP
Twenty-five sugarcane leaves which showed mosaic, interveinal chlorotic spots and leaf stripe symptoms were collected from sugarcane fields in Nanning City, Guangxi Province.The total RNA was extracted and detected by RT-LAMP.For the detection, 2 µL SYBR Green I was added into the tube after the RT-LAMP reaction was completed.
2 Results and analysis
2.1 Reaction condition optimization for SCSMV detection by one-step RT-LAMP assay
To optimize the temperature for the RT-LAMP assay used for SCSMV detection, we compared the results from reactions performed at different temperatures, including 60 ℃ , 61 ℃ , 62 ℃ , 63 ℃ ,64 ℃ and 65 ℃ , by using RNA samples extracted from SCSMV-infected sugarcane leaves(Fig.1).The results revealed that reactions at 64 ℃ and 65 ℃for 60 min produced low concentrations of ladder-like products.In contrast, reactions performed from 60 ℃to 63 ℃ led to the production of a clear ladder-like pattern upon agarose gel analysis(Fig.1).

Fig.1 Optimization of reaction temperature for RT-LAMP
M: DL2000 marker.Lane 1: Negative control; Lane 2-7: RT-LAMP reaction temperatures at 60℃, 61℃, 62℃, 63℃, 64℃ and 65℃,respectively
2.2 Specificity of RT-LAMP detection of SCSMV
The total RNA extracted from sugarcane leaves that were only infected by SCSMV, SCMV or SrMV(all verified by RT-PCR)was analyzed by RT-LAMP.RT-LAMP amplification only produced ladder-like bands when RNA from leaves infected by SCSMV(Fig.2).No signals were detected in samples prepared from healthy leaves or from those infected by SCMV or SrMV.
2.3 Sensitivity of RT-LAMP amplification
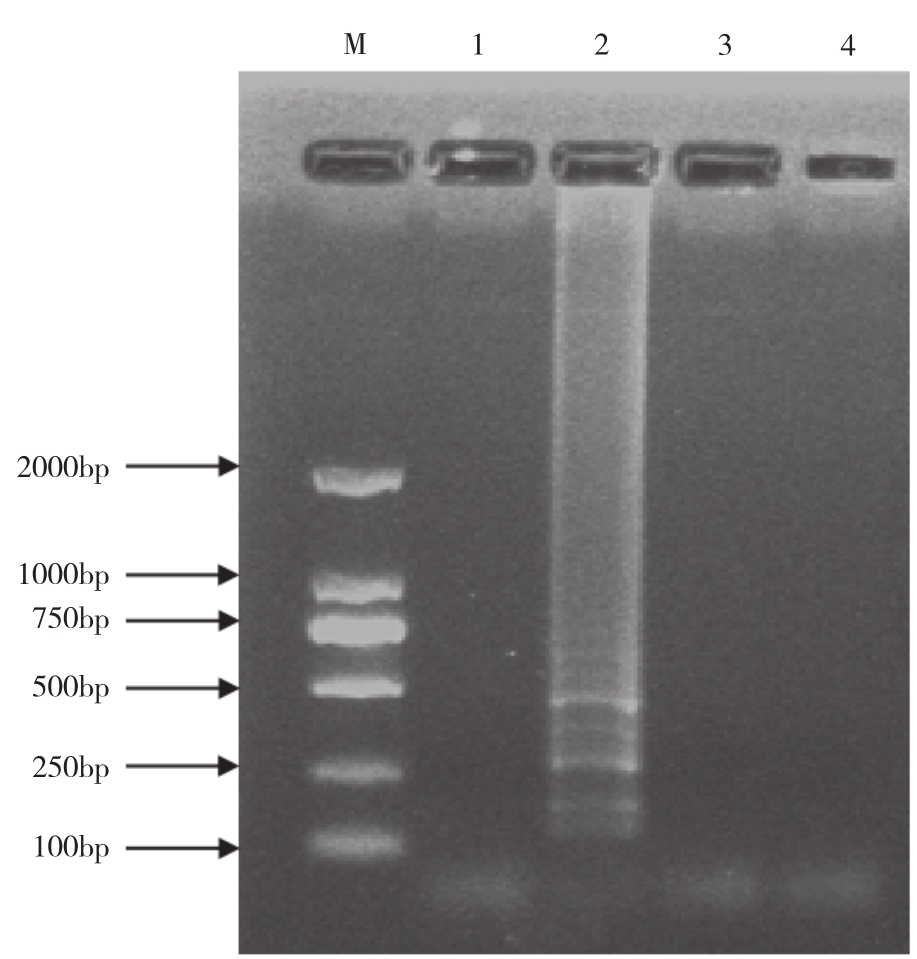
Fig.2 Specificity of RT-LAMP detection of SCSMV
M: DL2000 marker.Lane 1: Negative control; Lane 2-4: RT-LAMP detection of RNA extracted from SCSMV, SrMV infected sugarcane, respectively
To compare the sensitivity between the RTLAMP assay and RT-PCR, the assays were performed by using serial diluted templates, with the concentrations of RNA ranging from 167.9 ng to 167.9×10-8 ng.RT-LAMP amplification was able to generate clear DNA products when the template concentration was 167.9×10-5 ng(Fig.3A).At this template concentration, no product was detected in RT-PCR reactions.RT-PCR did not generate detectable DNA products except the template was used at 167.9×10-3 ng(Fig.3B).Therefore, under the reaction condition in this study, the RT-LAMP method is at least 100 times more sensitive than RT-PCR.
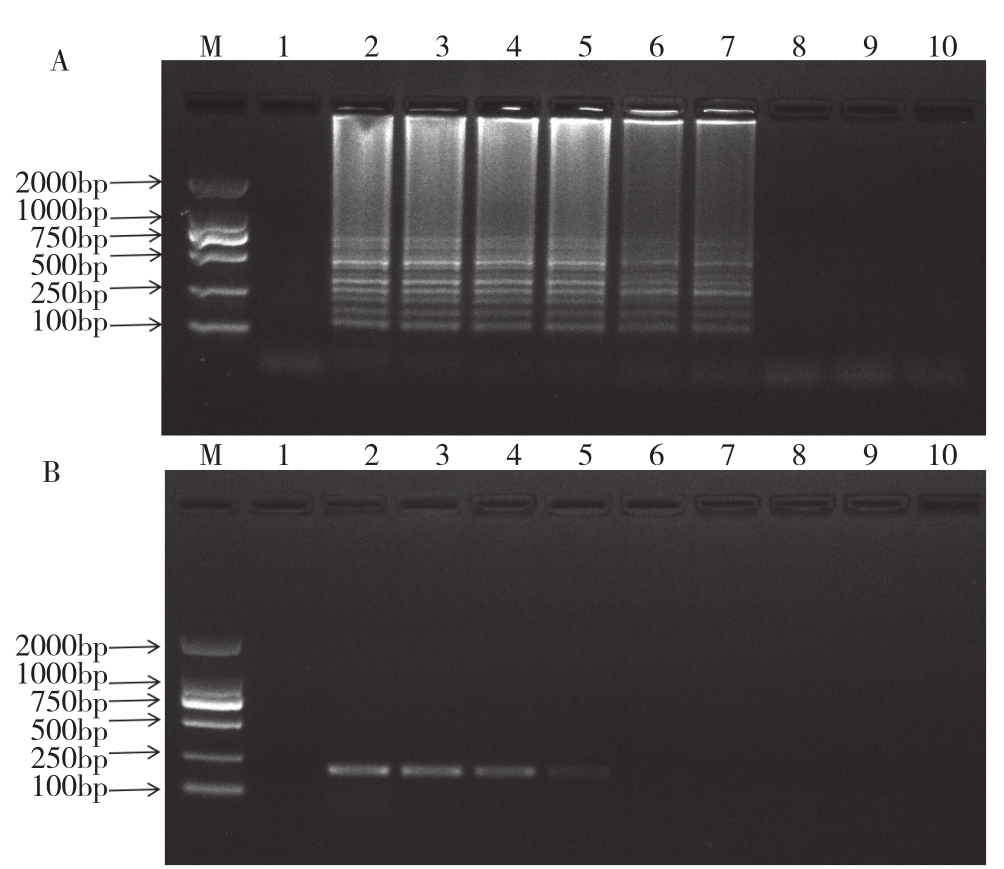
Fig.3 Comparison of sensitivity of RT-LAMP and RT-PCR for the detection of SCSMV
A: Sensitivity of RT-LAMP for the detection of SCSMV analyzed by agarose gel electrophoresis
B: Sensitivity of RT-PCR for the detection of SCSMV analyzed by agarose gel electrophoresis
M: DL2000 marker.Lane1: Negative control; Lane 2-10: Ten times dilution of total RNA(167.9 ng, 167.9×10-1 ng, 167.9×10-2 ng, 167.9×10-3 ng,167.9×10-4 ng, 167.9×10-5 ng, 167.9×10-6 ng, 167.9×10-7 ng, 167.9×10-8 ng)isolated from SCSMV infected sugarcane was included in each 25 µL(total volume)of RT-LAMP reaction
2.4 Detection of SCSMV in field samples by RTLAMP
To examine the usefulness of the RT-LAMP in detecting SCSMV in field samples, twenty-five leaf samples were collected from sugarcane that displayed typical mosaic symptoms from fields in Nanning City.The total RNA extracted was subjected to RTLAMP.Reactions with RNAs from five of the samples produced a clear ladder-like pattern after agarose gel analysis(partial results shown Fig.4A).To further simplify the detection procedure, 10 μL of the completed reaction was transferred to a test tube and a drop of SYBR Green I was added, allowing direct visualization of the RT-LAMP products by naked eyes.As shown in Fig.4B, the fluorescence dye of the RTLAMP reaction containing SCSMV infected samples changed from red to yellow, allowing direct visual detection.No color change occurred in tubes in which the samples detected negative for SCSMV.

Fig.4 RT-LAMP detection of partial field samples
A: RT-LAMP detection of partial field samples analyzed by agarose gel electrophoresis.M: DL2000marker; Lane1: Negative control; Lane 2: Positive control; Lane 3-7: Sugarcane samples collected from sugarcane fields
B: RT-LAMP detection of SCSMV analyzed by color changes.Tube1:Negative control; Tube 2: Positive control; Tube 3-7: sugarcane samples collected from sugarcane fields
3 Discussion
Sugarcane is an important economic crop in southern China, especially in Guangxi Province.Sugarcane mosaic disease, which caused by SCMV,SrMV and SCSMV, causes significant yield losses every year.Though breeding and cultivating resistant cultivars remain the most economical and effective strategy, accurate and early disease diagnosis in the laboratory and the field is desirable.Relying on its simple, rapid, specific and sensitive characteristics,RT-LAMP facilitate the simplicity of field use.RTLAMP assay has been used for detection of sugarcane samples infected by SCMV, SrMV, ratoon stunting disease(RSD), SCSMV[22,27-28].
In this study, specific primers have been designed corresponding to the conserved regions of the viral coat protein for SCSMV detection by using RT-LAMP.The primers were effective in detecting SCSMV, compared to conventional RT-PCR, the RT-LAMP assay established in this study showed higher sensitivity with 167.9×10-5 ng of infected sugarcane total RNA than that detected by RT-PCR with 167.9×10-3 ng.And the RT-LAMP assay showed high specificity, had no cross-reaction for other two mosaic viruses(SCMV and SrMV).The field samples were also detected by using the RT-LAMP assay, and the results were entirely consistent with the RT-PCR results.To further simplify the detection procedure, a SYBR Green I dye was also added in the test tube, and the result could be visualized by naked eyes.All of the results above showed that this method could be used for the detection of SCSMV.
The sensitivity of RT-LAMP may differ in the detection of different targets.Keizerweerd et al.[22]reported that the RT-LAMP assay was 10 times more sensitive than conventional RT-PCR in detecting SCMV and SrMV.And Le et al.[29] reported that the RT-LAMP assay was 100 times more sensitive than conventional RT-PCR in detecting nine rice viruses.In this study, the sensitivity of RT-LAMP for detection of SCSMV was 100 times more sensitive than conventional RT-PCR, similar as earlier reports of RT-LAMP for detection of other viruses[22,29], but higher than Wang’s report of RT-LAMP for detection of SCSMV[28].However, there were also report that the sensitivity of RT-LAMP for detection of Bean pod mottle virus was at least 1 000 times more sensitive than conventional RT-PCR[30].The length of the primer,the quality of the primer, annealing temperature and GC contents could infect the sensitivity of the RTLAMP amplification.Changing the location of primers may improve the sensitivity of detection.For example,after changing the location of primers, the sensitivity of RT-LAMP assay was found higher in detecting SCSMV in this study than that of Wang et al.[28].Therefore, in order to improve the sensitivity of RTLAMP detection, changing the location of primers may be a good attempt.
4 Conclusion
In conclusion, a one-step RT-LAMP assay was developed as an alternative method for the rapid detection of SCSMV.The whole process can be completed in 60 minutes at 60-63℃.The RT-LAMP assay had no cross-reaction for other two mosaic viruses(SCMV and SrMV)and showed high sensitivity, 100 times more than conventional RT-PCR.Field samples can also be accurately detected by RT-LAMP assay established in this study.Therefore, this assay could be used for health management program and surveillance of sugarcane disease in laboratories or the detection of SCSMV in the field to prevent the disease spread.
[1] HEMA M, SREENIVASULU P, SAVITHRI H S.Taxonomic position of Sugarcane streak mosaic virus in the family Potyviridae[J].Archives of Virology, 2002, 147(10):1997–2007.doi:10.1007/s00705-002-0851-1.
[2] LI W F, HE Z, LI S F, HUANG Y K, ZHANG Z X, JIANG D M, WANG X Y, LUO Z M.Molecular characterization of a new strain of Sugarcane streak mosaic virus(SCSMV)[J].Archives of Virology, 2011,156(11):2101-2104.doi:10.1007/s00705-011- 1090-0.
[3] HALL J S, ADAMAS B, PARSONS T J, FRENCH R, LANE L C,JENSEN S G.Molecular cloning, sequencing, and phylogenetic relationships of a new potyvirus: Sugarcane streak mosaic virus, and a reevaluation of the classification of the potyviridae[J].Molecular Phylogenetics and Evolution, 1998,10(3):323–332.
[4] HEMA M, IRTHI N, SREENIVASULU P, SAVITHRI H S.Development of recombinant coat protein antibody based IC-RTPCR for detection and discrimination of Sugarcane streak mosaic virus isolates from Southern India[J].Archives of Virology, 2003,148(6):1185-1193.doi:10.1007/s00705-003-0015-y.
[5] WYLIE SJ, TAN A J Y, LI H, DIXON KW, JONES M G K.Caladenia virus A, an unusual new member of the family Potyviridae from terrestrial orchids in Western Australia[J].Archives of Virology,2012,157(12): 2447–2452.doi:10.1007/s00705-012-1452-2.
[6] PARAMESWARI B, BAGYALAKSHMI K, VISWANATHAN R,CHINNARAJA C.Molecular characterization of Indian Sugarcane streak mosaic virus isolate[J].Virus Genes, 2013, 46(1):186–189.doi:10.1007/s11262-012-0827-5.
[7] CHATENT M, MAZARIN C, GIRARD J C, FERNANDEZ E,GARGANI D, RAO G P, ROYER M, LOCKHART B, ROTT P.Detection of Sugarcane streak mosaic virus in sugarcane from several Asian countries[J].International Society of Sugar Cane Technologists Proceedings of the XXV Congress, 2005,23:12-15,28.
[8] DAMAYANTI T A, PUTRA L K.First occurrence of Sugarcane streak mosaic virus infecting sugarcane in Indonesia[J].Journal of General Plant Pahtology, 2011,77(1):72–74.doi:10.1007/s10327-010-0285-7.
[9] PUTRA L K, KRISTINI A, ACHADIAN E M, DAMAYANTI T A.Sugarcane streak mosaic virus in Indonesia: distribution,characterisation, yield losses and management approaches[J].Sugar Tech, 2014,16(4):392–399.doi:10.1007/s12355-013-0279-9.
[10] VISWANATHAN R, BALAMURALIKRISHNAN M, KARUPPAIAH R.Characterization and genetic diversity of Sugarcane streak mosaic virus causing mosaic in sugarcane[J].Virus Genes, 2008,36(3):553–564.doi:10.1007/s11262-008-0228-y.
[11] XU D L, ZHOU G H, XIE Y J, Mock R, LI R.Complete nucleotide sequence and taxonomy of Sugarcane streak mosaic virus, member of a novel genus in the family Potyviridae[J].Virus Genes, 2010,40(3):432-439.doi:10.1007/s11262-010-0457-8.
[12] HE Z, LI W F, YASAKA R, HUANG Y K, ZHANG Z X, OHSHIMA K, LI S F.Molecular variability of Sugarcane streak mosaic virus in China based on an analysis of the P1 and CP protein coding regions[J].Archives of Virology, 2014,159(5): 1149-1154.doi:10.1007/s00705-013-1854-9.
[13] HE Z, YASAKA R, LI W F, LI S F, OHSHIMA K.Genetic structure of populations of Sugarcane streak mosaic virus in China: Comparison with the populations in India[J].Virus Reseach, 2016,211: 103-116.doi:10.1016/j.virusres.2015.09.020.
[14] REDDY C V S, SREENIVASULU P, SEKHAR G.Dupleximmunocapture-RT-PCR for detection and discrimination of two distinct potyviruses naturally infecting sugarcane(Saccharum spp.hybrid)[J].Indian Journal of Experimental Biology, 2011,49(1):68–73.
[15] SINGH D, TEWARI A K, RAO G P, KARUPPAIAH R,VISWANATHAN R, ARYA M, BARANWAL VK.RT-PCR/PCR analysis detected mixed infection of DNA and RNA viruses infecting sugarcane crops in different states of India[J].Sugar Tech, 2009,11(4):373–380.doi:10.1007/s12355-009-0064-y.
[16] PRABOWO D B, HADIASTONO T, HIMAWAN T, PUTRA L K.Detection disease of sugarcane streak mosaic virus(scsmv)via serological test on sugarcane(Saccharum officinarum L.)[J],International Journal of Science and Research, 2014,3(1): 88–92.
[17] FENG X Y,SHEN L B,WANG W Z,WANG J G,CAO Z Y,ZHANG S Z.Reverse transcription–recombinase polymerase amplification assay for the detection of sugarcane streak mosaic virus in sugarcane[J].Sugar Tech,2019,21(4):645-652.doi:10.1007/s12355-019-0051-9.
[18] FU W L, SUN S R, FU H Y, CHEN R K, SU J W, GAO S J.A one-step real-time rt-pcr assay for the detection and quantitation of sugarcane streak mosaic virus[J].Biomed Research International, 2015.doi:10.1155/2015/569131.
[19] HEMA M, VENKATRAMANA M, SAVITHRI H S, SREENIVASULU P.Biological, antigenic and genomic relationships among the virus isolates causing mosaic disease of sugarcane in South India[J].Current Science, 1999,77(5): 698-702.
[20] ZHANG K, CHEN C F, XU H M, CHEN J H,CHEN W, HE Z.Rapid and Sensitive Identification of the SCSMV-infected sugarcane based on immuno-detections[J].Acta phytopathologica sinica, 2019(online)1-18.doi:10.13926/j.cnki.apps.000423.
[21] NOTOMI T, OKAYAMA H, MASUBUCHI H, YONEKAWA T,WATANABE K, AMINO N, HASE T.Loop-mediated isothermal amplification of DNA[J].Nucleic Acids Research, 2000,28(12):E63.doi:10.1006/mpev.1998.0535.
[22] KEIZERWEERD A T, CHANDRA A, GRISHAM M P.Development of a reverse transcription loop-mediated isothermal amplification(RT-LAMP)assay for the detection of Sugarcane mosaic virus and Sorghum mosaic virus in sugarcane[J].Journal of Virological Methods,2015,212: 23-29.doi:10.1016/j.jviromet.2014.10.013.
[23] LIU Z M, XIA X Y, YANG C Y, HUANG J Y.Colorimetric detection of Maize chlorotic mottle virus by reverse transcription loop-mediated isothermal amplification(RT-LAMP)with hydroxynapthol blue dye[J].RSC Advances, 2016,6(1): 73–78.doi:10.1039/C5RA20789D.
[24] WANG Z Y, GU Q S, SUN H, LI H L, SUN B J, LIANG X Z, YUAN Y, LIU R L, SHI Y.One-step reverse transcription loop mediated isothermal amplification assay for sensitive and rapid detection of Cucurbit chlorotic yellows virus[J].Journal of Virological Methods,2014,195: 63–66.doi:10.1016/j.jviromet.2013.08.037.
[25] XU D L, ZHOU G H, SHEN W K, DENG H H.Genetic Diversity of Sorghum mosaic virus infecting sugarcane[J].Acta Agronomica Sinica,2008,34(11): 1916-1920.
[26] XU D L, ZHOU G H.Genetic diversity of SCMV infecting sugarcane in South China[J].Acta Phytopathology Sinica, 2005,35(6):143-144.doi:10.13926/j.cnki.apps.2005.s1.037.
[27] GHAI M,SIGH L A,MARTIN L, MCFARLANE S A, VAN T,RUTHERFORD R S.A rapid and visual loop-mediated isothermal amplification assay to detect Leifsonia xyli subsp. xyli targeting a transposase gene[J].Letters in Applied Microbiology, 2013,59(6):648-657.doi.org/10.1111/lam.12327.
[28] WANG K L,DENG Q Q,CHEN J W,SHEN W K.Development of a reverse transcription loop-mediated isothermal amplification assay for rapid and visual detection of Sugarcane streak mosaic virus in sugarcane[J].Crop Protection, 2019,119:38-45.doi:10.1016/j.cropro.2018.11.024.
[29] LE D T, NETSU O, UEHARA-ICHIKI T, SHIMIZU T, CHOI I R,OMURA T, SASAYA T.Molecular detection of nine rice viruses by a reverse-transcription loop-mediated isothermal amplification assay[J].Journal of Virological Methods, 2010,170(1/2):90-93.doi.org/10.1016/j.jviromet.2010.09.004.
[30] WEN W G,YANG C Y, CAI J X,ZHANG Y.Detection of Bean pod mottle virus by RT-LAMP[J].Plant Protection, 2010,36(6):139-141.