文章信息
基金项目
- 海南省自然科学基金(318QN189);海南省教育厅项目(Hnky2021-19,Qhys2021-248)
作者简介
- 周扬(1988—),男,博士,副教授,研究方向为植物抗逆机理,E-mail:zhouyang@haiananu.edu.cn.
文章历史
- 收稿日期:2023-08-29
土壤盐渍化是指高浓度可溶性盐分(如Na+、Cl-、SO42-、HCO3-等)在土壤中累积导致土壤性质恶化和生产质量下降的过程[1]。据联合国粮农组织2021年发布的全球盐渍土壤分布图显示,全球盐渍土壤面积逾8.33亿hm2,占地球面积的8.7%。中国盐渍土面积近1亿hm2,从热带到寒温带、滨海到内陆、湿润到极端干旱的地区均有分布[2],是继澳大利亚和俄罗斯之后世界第三大盐碱地分布区[3]。与此同时,农业生产中化肥滥用以及不合理的灌溉方式导致了大量农田发生次生盐渍化[4]。根据耐盐性不同,可将植物分为甜土植物(Glycophytes)和盐生植物(Halophytes)[5]。大多数的农作物(如水稻、小麦和玉米等)均属甜土植物,当土壤中的NaCl含量超过200 mmol/L时引发植物死亡,有些园艺作物抵抗盐胁迫的能力更低[6-7]。2022年中央一号文件指出要“分类改造盐碱地,推动由主要治理盐碱地适应作物向更多选育耐盐碱植物适应盐碱地转变”。因此,推动“以种适地”同“以地适种”相结合,加快选育耐盐碱作物品种,才能充分挖掘盐碱地综合利用潜力,提高农业综合生产能力。植物的耐盐性是一个由多基因控制的复杂的生物学性状,目前研究人员已经发现多个在盐胁迫信号转导中起作用的关键基因及调控通路。2023年8月农业农村部指出“以转基因为代表的生物育种是育种领域的革命性技术,是必须抢占的新领域新赛道”。因此,利用现代分子育种手段培育耐盐作物品种是未来育种工作的重点。本文根据基因在细胞中的位置不同,将其分为细胞壁、细胞质膜、细胞内膜系统上的耐盐基因,对其耐盐性及调控机制进行综述,以期为利用分子生物学技术加快选育耐盐作物新品种提供理论基础。
1 植物细胞壁系统上的耐盐基因植物细胞壁结构的完整性对于维持其生长和抵抗胁迫起重要作用[8]。植物在生长发育过程中不断感知和响应外界环境因子,细胞壁上的类受体激酶(Receptor-like kinase,RLK)可识别激素、小肽、糖类等信号分子,以适应外界环境。快速碱性化因子(Rapid alkalinization factor,RALF)是一种肽类激素,作为受体激酶CrRLK1L(Catharanthus roseus RLK1-like kinase)亚家族的配体,二者形成的复合体CrRLK1L-RALF在植物细胞中的多个生物学过程中起调控作用[9]。拟南芥RALF1通过与其受体CrRLK1L家族成员Feronia(FER)相互作用调节植物生长[10],RALF23与FER相互作用调节植物免疫[11]。盐胁迫下,定位于细胞壁上的富含亮氨酸重复序列的伸展蛋白(Leucine-rich repeat extensins,LRX)通过其LRR结构域与FER-RALF22/23复合体结合,形成的LRXs-RALFs-FER蛋白复合物可协同调控植物耐盐性[12]。类受体激酶RLK7的胞外结构域与PAMP-INDUCED SECRETED PEPTIDE3(PIP3)小肽结合,二者均受盐胁迫显著诱导,PIP3-RLK7模块可以启动下游信号调控网络,提高植物耐盐性[13]。
细胞壁的主要成分纤维素在植物响应盐胁迫中有着重要作用,纤维素合酶CESA6(Cellulose synthase 6)能够与其互作蛋白CSI1(Cellulose synthase-interactive protein 1)发生作用,参与调控拟南芥耐盐性[14]。当植物处于高盐环境时,纤维素合酶复合物的两个组成部分CC1(Companions of cellulose synthase,CC)和CC2的活性增加,结合并支撑微管组织,调控盐胁迫下微管的解聚和重建,同时使纤维素合酶定位在质膜上,调控纤维素的合成,保证植物细胞壁的防御功能,最终使幼苗对盐胁迫不敏感[15]。当植物受到盐胁迫时,细胞壁上的纤维素生物合成受阻,细胞壁结构完整性受到破坏,这一信号会激活细胞表面的富含亮氨酸的重复受体激酶MIK2/LRR-KISS(Male discoverer 1-interacting receptor like kinase 2/Leucine-rich repeat kinase family protein induced by salt stress),MIK2参与感知植物细胞壁的扰动,调节根尖的细胞壁结构,从而保护植物免遭盐胁迫侵害[16]。MIK2与细胞壁完整性传感器THESEUS1(THE1)具有重叠和不同的功能,mik2突变体的盐胁迫敏感性表型依赖于THE1[17],说明MIK2以依赖THE1的方式参与纤维素生物合成抑制时引发的细胞壁损伤反应的响应[16]。
β-1,4-半乳聚糖是构成细胞壁果胶多糖的重要组分,在植物响应盐胁迫中起作用。盐胁迫下,拟南芥根部半乳聚糖合成酶1(GALACTAN SYNTHASE 1,GALS1)活性增强,进而提高细胞壁β-1,4-半乳聚糖含量,影响纤维素的合成,最终加强植物对盐胁迫的敏感性。进一步研究发现,GALS1的表达受转录因子BPC1和BPC2(Barleybrecombinant/Basicpentacysteine transcription factors)直接调控[18]。最新研究指出,盐胁迫激活的CBF1/CBF2/CBF3转录因子能够增强其与GALS1启动子的结合能力,直接识别并抑制GALS1表达,导致半乳聚糖含量减少,缓解拟南芥的盐敏感性。同时,盐胁迫激活的CBF1/CBF2/CBF3以不依赖于BPC1/BPC2的方式对盐胁迫上调的GALS1表达施加“限制”,实现对GALS1的精准调控,有助于植物适应盐胁迫[19]。
植物细胞壁的另一种主要成分木质素也在盐胁迫响应中起作用。盐胁迫会刺激植物根系细胞壁的木质化,能够有效阻止根系对外界环境中盐离子的吸收,提高植物细胞的耐盐性[20]。Li等[21]对滨海盐生植物海马齿的耐盐分子机制进行研究,发现木质素在海马齿根部的积累是其抵御盐胁迫的一种适应性机制。结合转录组和代谢组测序,发现木质素合成途径中的肉桂酰辅酶A还原酶基因(CCR)、反式肉桂酸4-单加氧酶基因(CYP73A)和4-香豆酰基喹酸酯(香豆酰基莽草酸酯)3’-单加氧酶基因(C3’H)、咖啡酸3-O-甲基转移酶基因(COMT)、咖啡酰辅酶A-O-甲基转移酶基因(CCoAOMT)等在盐处理后表达量上升,木质素合成单体对香豆醇和芥子醇的含量增加,最终导致根系在盐胁迫下积累更多的木质素。
此外,细胞壁中的其他组分也参与到植物盐胁迫响应中。L-阿拉伯糖(Ara)是植物细胞果胶壁的主要成分,有呋喃糖(Araf)和吡喃糖(Arap)两种形式。UDP-D-木糖-4-差向异构酶1(UXE1)在Araf和Arap的转化过程中起重要作用。UXE1基因突变后的拟南芥突变体(MUR4)对盐胁迫敏感,根伸长表现异常,当外源施加Ara时,可恢复此突变表型;而当Ara含量降低时,拟南芥的盐敏感性加强,表明细胞壁中的阿拉伯糖代谢过程在植物抗盐过程中起重要作用[22]。
2 植物细胞膜系统上的耐盐基因 2.1 细胞膜通道蛋白调控耐盐性土壤中的盐离子随水通过共质体途径进入根系细胞,由于根系内皮层凯氏带的阻碍[23-24],盐进入植物根系需要一些通道蛋白或者转运体蛋白的协助。外界环境中的Na+可能通过细胞膜上的高亲和性K+通道(High-affinity K+ channel,HKT)、低亲和性K+通道(如Arabidopsis K+ Transporter 1,AKT1)、弱电压依赖性非选择性阳离子通道(Nonselective cation channel,NSCC)、电压非依赖性通道(Voltage-independent channel,VIC)和非选择性外流式电导(Nonselective outward-rectifying conductance,NORC)等途径进入细胞[25-27]。所有这些通道均属于离子通道,可介导Na+和K+流入植物细胞,且部分通道对K+的选择性比Na+强[25-26]。近期研究发现细胞膜上的GIPC鞘脂(Glycosyl inositol phosphorylceramide)是非离子通道盐受体,盐胁迫下外界环境中的Na+与GIPC结合,引起细胞表面电势变化,形成Ca2+内流通道,导致细胞质中的Ca2+浓度增加,产生钙信号以激活下游调控通路应对盐胁迫[28]。盐受体的发现说明质膜上的脂类可能参与了盐胁迫响应,未来可用于提高作物耐盐性。
细胞膜K+通道蛋白在植物耐盐调节中的作用机制研究较为清楚。当拟南芥处于高盐胁迫时,根系会在2 min内快速积累大量的Na+[29]。由于Na+和K+共享运输系统,因此盐胁迫下细胞吸收了过量Na+可能会抑制植物细胞对K+的吸收,从而导致植物K+缺乏,产生营养胁迫[30]。K+能够维持细胞代谢,细胞内酶的活动主要依靠K+的浓度,由于Na+与K+属于同族化学元素,在特征特性上具有相似性,盐胁迫下Na+在细胞质中积累导致K+被Na+取代,但Na+不能取代K+的功能。细胞质中高浓度的Na+破坏了必需的代谢途径,导致植物遭受严重的离子胁迫[25]。因此,植物在遭受盐胁迫时,可通过调节细胞膜上通道蛋白的活性来限制Na+进入细胞,以提高耐盐性[24]。
HKT转运蛋白是一种高亲和K+转运蛋白,但其也可转运Na+,帮助植物更好地完成吸K+排Na+,以维持细胞内的Na+-K+平衡[31]。HKT型转运体的耐盐功能首先是在拟南芥盐敏感突变体中发现的,研究发现拟南芥AtHKT1介导Na+在根中的内流[32-33],且具有单一的Na+选择性运输活性[34]。AtHKT1基因发生突变会导致茎中Na+过度积累和对Na+过度敏感,说明AtHKT1参与了Na+通过韧皮部从茎部向根的再循环[35]。与拟南芥中只含有一个HKT1基因不同,水稻、小麦和大麦等单子叶植物编码多个HKT型转运蛋白[36]。水稻有7个功能性的HKT型转运蛋白,根据转运活性可分为Ⅰ型和Ⅱ型。Ⅰ型HKT转运体通常介导Na+选择性转运;而Ⅱ型HKT转运体主要是Na+/K+转运体,共同运输Na+和K+[36-37]。小麦Ⅱ型HKT转运蛋白TaHKT2;1可同时运输Na+和K+,并且其转运K+的活性可被Na+激活[38-39]。Ⅰ型HKT转运蛋白是进化上重要的耐盐决定因子,其自然变异与耐盐性有关[40]。水稻OsHKT1;5可从根木质部去除Na+,从而提高其耐盐性[41-42]。一粒小麦(Triticum monococcum L.)TmHKT1;5-A以及普通小麦(Triticum aestivum L.)TaHKT1;5-D1与其拟南芥同源蛋白AtHKT1具有相似的功能,均可从根木质部卸载Na+[43]。将TmHKT1;5-A基因转化到硬粒小麦(Triticum turgidum ssp. Durum var. Tamaroi)中,在盐碱地培养时,籽粒产量提高了25%[44],表明该基因在提高作物耐盐性方面具有潜在的应用价值。
AKT1属于内向整流型钾离子通道(Inward rectifier K+ channel)[45],对钾离子的亲和力低、对钾离子浓度变化敏感、对电压的依赖性高。AKT1在调节植物体内K+/Na+平衡以及提高耐盐性等方面具有非常重要的作用。盐胁迫下,番茄AKT1可以维持细胞内K+/Na+的动态平衡,参与盐胁迫应答[46]。水稻OsAKT1可以控制根部对Na+的吸收来维持植株内的K+/Na+平衡[47]。盐胁迫下盐地碱蓬(Suaeda salsa)SsAKT1可促进植物体内K+累积而增强耐盐性[48]。在拟南芥中过表达小花碱茅(Puccinellia tenuiflora)PtAKT1基因后,其体内的K+积累增加、耐盐性提高[49]。在小麦中的研究发现,盐胁迫下耐盐品种中的 AKT1基因表达上调,而盐敏感品种该基因表达下调[50],暗示AKT1与小麦的耐盐性相关。
当土壤盐度过高时,NSCC是根吸收Na+的主要途径[51]。谷氨酸受体(Glutamate Receptors,GLRs)和环核苷酸门控通道(Cyclic Nucleotide-Gated Channels,CNGC)这两种NSCC参与根Na+摄取[52]。研究发现NSCC/VIC在植物根系皮层中介导Na+通过细胞膜进入植物体,NSCC受盐诱导信号cGMP和ROS的调控[53],抑制NSCC可缓解盐胁迫对植物造成的伤害,干扰VIC的活性可使植物体内积累更少的Na+[54]。
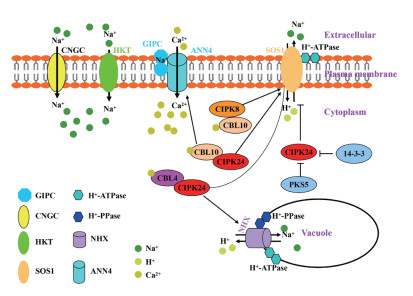
2.2 细胞膜Na+/H+逆转运蛋白参与钙信号途径调控耐盐性SOS1(Salt overly sensitive 1)作为目前唯一已知的细胞膜Na+外排蛋白,当外界环境中的Na+进入到细胞后,SOS1可在细胞膜上进行Na+/H+离子交换,将细胞质中过多的Na+外排。SOS1基因在拟南芥盐敏感突变体中被首先鉴定[55-56],其控制离子稳态中的作用已通过遗传、生化和生理等手段得到证明[57-59]。盐胁迫下,水稻、小麦和拟南芥中SOS1基因的表达量均增加[60-62]。生化分析表明,SOS1主要在根尖表皮细胞和根、茎、叶木质部的薄壁细胞中表达。在轻度胁迫条件下(25 mmol/L NaCl),sos1突变体地上部分Na+积累量低于野生型,而在重度胁迫条件下(100 mmol/L NaCl),突变体地上部分Na+积累量明显高于野生型[58]。以上结果表明,SOS1在Na+的长距离转运中也起着重要作用。
SOS途径由Na+/H+逆转运蛋白SOS1、丝氨酸/苏氨酸蛋白激酶SOS2(Calcineurin B-like interacting protein kinase 24,CIPK24)和钙结合蛋白SOS3(Calcineurin B-like 4,CBL4)组成(图 1),已在拟南芥[59, 63]、水稻[61]、杨树[64]等甜土植物,以及盐芥(Thellungiella salsuginea)[65]、海马齿[66]等盐生植物中被发现。盐胁迫下,拟南芥根部AtCBL4(AtSOS3)识别感知钙信号,作用于AtCIPK24(AtSOS2)的NAF/FISL结构域,形成的复合体磷酸化调控SOS1蛋白C-末端1 136和1 138位置的丝氨酸位点,增强SOS1的活性[59]。类似于SOS2的蛋白激酶(SOS2-like protein kinase 5,PKS5)可以与SOS2相互作用并磷酸化SOS2的第294位氨基酸,促进SOS2与14-3-3蛋白的相互作用,抑制SOS2活性,从而负向调控SOS途径[67]。盐胁迫下的钙信号是由14-3-3蛋白和SOS3蛋白共同解码,二者选择性地激活/失活下游蛋白激酶SOS2和PKS5,通过协调介导SOS1的活性来调节Na+稳态[67]。盐胁迫下,AtANN4蛋白发挥Ca2+通道活性,内向转运Ca2+,参与激活SOS途径以应对盐胁迫。这一过程受到SCaBP8(SOS3-like calcium binding protein 8)的介导,在SCaBP8作用下,AtANN4与SOS2互作并增强SOS2对AtANN4的磷酸化修饰,形成稳定的SCaBP8-AtANN4-SOS2蛋白复合体,参与植物长期的盐胁迫响应[68]。此外,SOS2还可磷酸化修饰CONSTITUTIVE TRIPLE RESPONSE1(CTR1)的激酶活性,SOS2-CTR1模块能够激活乙烯信号反应,导致植物抗盐性增强[69]。
|
| 图 1 植物细胞内Na+吸收、外排和区隔化模式 Fig. 1 Model of uptake, exclusion and compartmentation of Na+ in plant cells |
SOS1蛋白N端由12~13个跨膜结构域组成[70],C端的细胞质区域在功能上可分为功能激活区和自抑制区,中间由连接区连接,激活区和自抑制区之间的相互作用可调节SOS1蛋白的Na+/H+逆转运活性[59]。在正常条件下,拟南芥SOS1会形成同型二聚体,SOS1被锁定在封闭状态,从而屏蔽Na+/H+结合位点[71],维持低水平的Na+/H+逆转运活性[60, 72-73]。当盐胁迫引起的钙信号被感知时,SOS2蛋白激酶及SOS2-SOS3复合体会识别并磷酸化SOS1自抑制结构域1 136和1 138位的丝氨酸,解除SOS1的自抑制作用,从而激活SOS1[59, 74];当该位点丝氨酸突变为丙氨酸时,SOS1不能被磷酸化,进而降低了拟南芥对盐胁迫的耐受性[59, 63]。把1 136和1 138位置上的丝氨酸残基突变为天冬氨酸残基用于模拟磷酸化,可增强与SOS2的结合[72]。研究还发现,SOS1蛋白功能域下游的自抑制区域缺失突变后,其Na+/H+逆转运活性被组成性激活,表现出超高的耐盐性[75-77]。在烟草中过表达小麦TaSOS1的超活性突变体TaSOS1-974后,在250 mmol/L NaCl处理下,转TaSOS1-974烟草的苗高和鲜重与对照相比分别降低21% 和41%,而转全长基因TaSOS1的烟草苗高和鲜重与对照相比分别降低37% 和52%[77]。
植物细胞中的钙结合蛋白CBL感知由环境刺激产生的钙信号后,能够与CIPK蛋白互作来应对环境胁迫,构成了植物体特有的CBL-CIPK信号途径[78]。CBL4-CIPK24调控SOS1耐盐性除了在模式植物拟南芥中被发现外,在其他物种如水稻、杨树[61]、苹果[79]和番茄[80]中也被[81]鉴定。AtCIPK24还可与AtCBL10互作,在植物地上部参与调节AtSOS1蛋白活性[82]。AtCBL4定位于质膜,而AtCBL10位于液泡膜,CBL4和CBL10基因在异位突变体中表达后不能恢复其在盐胁迫下的表型[82-83],表明CBL蛋白在细胞中的位置决定了CBL-CIPK复合物的特异性生物学功能。Yin等[63]发现拟南芥AtCBL10能够与AtCIPK8互作于细胞膜上,调节SOS1的耐盐性(图 1)。海马齿中也发现存在一条耐盐通路SpCBL10-SpCIPK8-SpSOS1,过表达海马齿SOS1/CIPK8/CBL10可显著增强转基因酵母耐盐性[66]。
3 植物内膜系统的耐盐基因高等植物细胞中含有一个中央大液泡。植物通过Na+/H+交换在液泡中积累Na+[84],减少胞质中的Na+含量,从而缓解盐胁迫造成的危害。在液泡中积累Na+也可降低细胞水势、促进水分吸收。此外,在根系细胞液泡中积累Na+可减少Na+在叶片中的积累,降低Na+对叶片的损伤,保证叶片光合作用正常进行。更重要的是,在植物生长过程中细胞会产生更多的液泡空间,因此在液泡中存储Na+是减缓Na+毒性的有效机制[85]。
液泡膜上的Na+/H+交换活性最早是在拟南芥中发现的[86],该过程由Na+/H+离子交换器(Na+/H+ exchanger,NHX)完成。过表达拟南芥AtNHX1可提高多种植物如拟南芥、番茄、油菜等的耐盐性[40, 87-89]。在拟南芥中已鉴定出8个NHX基因,NHX1~NHX4蛋白位于液泡膜上,其中NHX1和NHX2是拟南芥中含量最多的蛋白[90],参与调控细胞内的pH和K+平衡[91];NHX5和NHX6蛋白位于细胞内膜系统如高尔基体膜(Golgi)、反式高尔基囊泡(Trans-Golgi vesicles)或者前液泡区(Prevacuolar compartments)中,主要参与调控细胞内的pH[92];NHX7和NHX8位于质膜中,NHX7即SOS1蛋白,主要负责细胞中的Na+外排[60],而NHX8主要介导Li+/H+的交换[93]。AtNHX1的拓扑结构和活性研究表明,AtNHX1的C端部分以Ca2+依赖的方式与液泡内的钙调蛋白(Calmodulin,CaM)相互作用,这种相互作用抑制AtNHX1的运输活性[94-95],当AtNHX1蛋白C末端的氨基酸位点发生突变后(P494L,P512S),可影响负调控因子CaM15与AtNHX1蛋白C端的结合,进而解除这一抑制作用[96]。综上,Na+通过Na+/H+逆转运蛋白进入液泡,是由Ca2+信号途径中的Ca2+结合蛋白CaM控制的。
NHX逆转运Na+是由2种不同类型的H+泵(H+-ATPase和H+-PPase)产生的质子(H+)电化学梯度驱动(图 1)[97]。在水稻中过量表达水稻液泡膜Na+/H+逆转运蛋白基因OsNHX1和液泡膜H+-焦磷酸酶基因OsVP1,水稻幼苗经100 mmol/L NaCl处理2周后,仍能存活,而未转基因水稻存活率只有38.67%,转化2个基因的转基因植株在盐胁迫下的植株高度和鲜重均显著高于转单个基因,说明共转化OsNHX1和OsVP1可显著提高水稻耐盐性[98]。当在水稻中异源表达来自于盐生植物碱蓬的SsNHX1基因和来源于拟南芥的AtAVP1基因时,转基因水稻幼苗在含150 mmol/L NaCl的MS培养基上比SsNHX1转化植株和未转化对照生长得更好,通过测定共转化植株液泡膜囊泡的V-PPase水解酶活性,也发现共转化植株的酶活性更高,这一结果也证明SsNHX1和AVP1共表达比单个表达SsNHX1引起的植物耐盐性更强[99]。将小麦TaNHX1和H+-PPase基因TVP1在模式植物拟南芥和烟草中异源表达,发现共转化2个基因的植株耐盐性与单个基因的植株相比得到明显提高[100-101]。上述研究结果表明,NHX1蛋白可提高植物耐盐性,但在植物体内共转化耐盐调控途径中的多个基因,其耐盐性将会得到明显提高。Zhou等[102]对125份野生大豆的耐盐性进行分析,利用蛋白质组学分析耐盐品系和盐敏感品系,筛选到5个差异表达的液泡膜转运蛋白(GsTIP2-1、GsTIP2-2、GsINT1、GsSUC4和GsACA11),发现它们可通过减轻对蛋白质合成和氨基酸代谢的抑制,从而提高大豆盐耐受性。
CBL-CIPK信号途径除了参与调控细胞膜系统应对盐胁迫,还能调控液泡膜上的蛋白参与Na+运输。研究发现,SOS2可正调控液泡膜H+-ATPase的活性和Na+/H+交换活性,以降低细胞质中Na+的毒性[103-104]。CBL10与液泡膜上的SOS2相互作用[105],调控液泡膜上一个未知的Na+通道转运蛋白,表明CBL10参与Na+在液泡中的区隔化[27]。此外,CBL2和CBL3通过与CIPK21结合,将CIPK21招募到液泡膜上来调节拟南芥的耐盐性[106]。这些发现丰富了目前植物耐盐性的CBL-CIPK调控机制。
4 植物耐盐基因的转录调控转录是调控基因表达的第一步,也是最关键的一步。转录因子是一类具有特殊结构,通过结合目的基因启动子区域中的顺式作用元件,能够激活或抑制目的基因的转录,从而使其在特定的时间与空间进行表达[107]。对盐胁迫处理下的拟南芥早期转录组分析表明,盐胁迫会导致数百至数千个基因的差异表达[108-109]。迄今为止,在许多植物中已经进行了与盐胁迫相关的转录因子研究,发现所有主要转录因子家族的成员(如MYB、WRKY、NAC、bZIP和ERF/AP2等)均会参与盐胁迫响应。
R2R3型MYB转录因子在响应盐胁迫中起主要作用。盐应答MYB转录因子通过结合靶基因启动子中的MYBCORE等顺式作用元件,激活或抑制其时空表达,参与ABA或者MAPK信号转导途径。也有一些MYB转录因子通过非ABA依赖途径调节耐盐性。拟南芥AtMYB73转录因子是SOS途径的负调节因子,atmyb73突变体中AtSOS1和AtSOS3基因在盐胁迫下的表达量升高,植物表现出耐盐性[110]。在拟南芥中过量表达芝麻SiMYB75转录因子,引起ABA合成途径中的基因AtNCED3和AtABA3的表达量上调,导致ABA含量增加、促进气孔关闭、减少叶片水分损失,提高了拟南芥的耐盐性[111]。而菠萝AcoMYB4通过结合ABA合成途径中AcoABA1基因的启动子,减少ABA的合成,对盐胁迫响应产生负调控[112]。在白杨中研究发现,R2R3型MYB基因PtrSSR1的表达受盐胁迫诱导,过表达该基因的转基因拟南芥的耐盐性有所提高,同时ABA相关基因NCED3、ABI1和CBL1表达量也随之上调,推测PtrSSR1通过调控ABA信号通路提高转基因拟南芥的耐盐性[113]。
WRKY转录因子主要是与基因启动子中的W-box结合,通过促进或者抑制SOS途径基因的表达,从而正调控或者负调控SOS途径的盐应答反应。高粱SbWRKY50能通过降低拟南芥AtSOS1的表达量而负调控植株的耐盐性[114]。金柑FcWRKY40则可以直接激活SOS途径中的FcSOS2基因的表达,间接调控SOS途径中的FcSOS1和FcSOS3基因的表达,促进Na+外排,从而提高耐盐性[115]。盐应答NAC转录因子通过结合启动子NACRS顺式作用元件(CACG),参与调控乙烯、生长素和ABA信号转导途径。苹果MdNAC047通过诱导乙烯合成基因MdACS1和MdACO1以及转录因子MdERF3的上调表达,积累乙烯并通过调节乙烯反应来增强耐盐性[116]。大豆GmNAC109基因在拟南芥中过量表达时,一方面可以诱导生长素合成途径中的基因AtAIR3(类枯草杆菌蛋白酶)的上调表达,另一方面会抑制AtARF2转录因子的表达,最终促进转基因拟南芥侧根的形成,从而表现出对高盐胁迫的耐受性[117]。马铃薯NAC家族基因StNAC053的表达受盐胁迫和ABA诱导,过表达该基因后转基因植株的耐盐性有所增强,同时发现在外源ABA处理下,转基因植株种子的发芽率低于野生型,说明NAC转录因子可能通过ABA途径参与植物耐盐[118]。苦荞(Fagopyrum tataricum)FtbZIP83转录因子能与FtSnRK2.6/2.3蛋白激酶互作,转化拟南芥后会引起拟南芥AtRD29A、AtRD29B和AtAIL等ABA诱导基因的上调表达,说明二者互作调控植物耐盐性可能依赖于ABA途径[119]。此外,FtSnRK2.6还能与苦荞FtbZIP5互作,通过调控ABA信号途径,减轻了转基因拟南芥在盐胁迫下的氧化损伤[120]。近期研究发现,bHLH转录因子PIF1和PIF3可与光敏色素phyA和phyB互作,负调控光形态建成,PIF1和PIF3的活性受到SOS2的磷酸化调控,从而调控植物在光照条件下的耐盐性[121]。小麦BR信号核心转录因子BRASSINAZOLE-RESISTANT1(TaBZR1)在盐处理后显著上调,过表达TaBZR1的转基因小麦在盐胁迫下积累的MDA和H2O2更少,表明TaBZR1的过表达增强了小麦的ROS清除能力[122]。综上,转录因子在植物响应盐胁迫的转录调控方面具有非常重要的作用。
虽然目前已对转录因子的功能进行了研究,但大多数转录因子的具体作用机制尚不清楚。转录因子是高度动态的,其调控目的基因的转录网络在特定的空间和时间起作用。转录组、全长转录组、空间转录组等组学技术的发展将有助于科研人员更精确的鉴定参与盐胁迫响应的转录因子。因此,未来需借助组学技术深入分析转录因子的耐盐调控机理,以确定盐胁迫反应转录调控的关键因素。
5 结语与展望植物耐盐性是由复杂的基因调控网络决定的,植物对盐的感知和反应的调控网络已较为清晰。植物中多种耐盐性必需的基因被相继报道,这些基因可能是培育耐盐作物的重要候选基因。尽管目前对CBL和CIPK已做了很多详尽的研究,但仍有较多的CBL、CIPK体外功能未被揭示。现有研究表明CBL和CIPK互作而构成的转导Ca2+信号的调控体系是非常复杂的,由于CBL和CIPK都是以基因家族形式存在,且基因家族成员彼此之间有较高的同源性,CBL与CIPK会发生交叉互作的情况,二者互作可能产生正调控或负调控的作用[123]。另外,植物体内的Ca2+信号途径还包括CDPK(Calcium dependent protein kinase)或MAPK(Mitogen-activated protein kinase)等,它们与CBL-CIPK信号途径也会发生交叉作用[76]。因此,要更清楚地理解植物体内复杂的耐盐调节通路,只能依赖于对植物CBL和CIPK基因功能更加清晰的解析。运用反向遗传学技术对基因进行突变,可快速清晰地获得基因的功能。同时,应把CBL-CIPK、CDPK、MAPK级联系统等信号模式整合在一起进行系统研究,才能更清楚地阐述植物对非生物逆境的应答机制。相信随着分子生物学技术和分子育种手段的成熟与完善,深入解析CBL和CIPK基因的功能以及CBL-CIPK信号通路在植物耐盐调节中的作用,有效放大和应用各基因成员的功能,将为植物的耐盐育种作出贡献。
从以往的研究中可以看到,转化耐盐调控途径中的多个基因要比转化单个基因更加有效。目前的转基因育种通常将调节植物耐盐性的不同基因和突变功能基因结合起来,利用已知耐盐机制的遗传变异来提高作物产量[98-99, 124]。SOS信号通路在高等植物中普遍存在,通过表达外源SOS通路调控基因,可提高生物体的耐盐性。高羊茅中共表达拟南芥SOS途径中的SOS1/2/3基因的转基因表现出更强的耐盐性[125]。转运Na+是一个主动运输的过程,SOS1在细胞膜上交换Na+需要质膜H+-ATPase提供能量[126],拟南芥中过表达海马齿SpSOS1和SpAHA1基因表现出更高的耐盐性,盐胁迫下上述拟南芥鲜重比转化单个SpSOS1或者SpAHA1提高26% 和33%,而比野生型拟南芥提高67%[127]。过表达拟南芥AtSOS1超活性突变体AtSOS1-Δ998或SOS2T168D/Δ308的转基因酵母具有耐盐优势[59]。小麦TaSOS1-974和拟南芥AtSOS-Δ998不仅在转基因酵母细胞中表现出相似的耐盐性,而且在转基因植物中也具有明显的耐盐性[77, 128]。因此,可利用分子生物学手段过表达这些基因或者其超活性的突变基因,如AtSOS1-Δ998、AtSOS2T168D/Δ308、TaSOS1-974[89, 93],或共同表达SOS通路基因[125-127, 129-130],可能会产生出具有高耐盐性的作物品种。
然而,将获得的耐盐基因转化到植物中,是否会导致植物产量品质明显降低是一个需要考虑的问题。基因编辑技术对耐盐目标基因的定点改造可以作为未来分子育种工作的重点用以培育耐盐作物。此外,盐生植物经过长期的适应性进化,在体内已形成了一套有别于甜土植物的耐盐调控网络,从盐生植物中挖掘耐盐基因进行甜土植物的耐盐性遗传改良,或者直接对盐生植物进行驯化,可能是一种有效的耐盐育种策略。近来的研究发现,利用作物野生近缘种的遗传多样性,从头驯化,有望产生更优良的作物品种。野生植物具备丰富的遗传变异,是拓展作物品种创新空间的重要资源,从头驯化是一种新育种策略,创新性地将野生植物直接作为育种底盘,通过重新设计和直接导入驯化基因快速创制新作物。总之,全面掌握植物耐盐机制,挖掘耐盐途径的关键基因,对利用分子生物学技术培育耐盐作物具有重要的应用价值。
| [1] |
ZHANG Y T, HOU K, QIAN H, GAO Y Y, FANG Y, XIAO S, TANG S Q, ZHANG Q Y, QU W G, REN W H. Characterization of soil salinization and its driving factors in a typical irrigation area of northwest China[J]. Science of the Total Environment, 2022, 837: 155808. DOI:10.1016/j.scitotenv.2022.155808 |
| [2] |
杨劲松, 姚荣江, 王相平, 谢文萍, 张新, 朱伟, 张璐, 孙瑞娟. 中国盐渍土研究: 历程、现状与展望[J]. 土壤学报, 2022, 59(1): 10-27. DOI:10.11766/trxb202110270578 YANG J S, YAO R J, WANG X P, XIE W P, ZHANG X, ZHU W, ZHANG L, SUN R J. Research on salt-affected soils in China: History, status and prospect[J]. Acta Pedologica Sinica, 2022, 59(1): 10-27. DOI:10.11766/trxb202110270578 |
| [3] |
云雪雪, 陈雨生. 国际盐碱地开发动态及其对我国的启示[J]. 国土与自然资源研究, 2020, 1: 84-87. DOI:10.16202/j.cnki.tnrs.2020.01.020 YUN X X, CHEN Y S. International development of saline-alkali land and its enlightenment to China[J]. Territory & Natural Resources Study, 2020, 1: 84-87. DOI:10.16202/j.cnki.tnrs.2020.01.020 |
| [4] |
PARK H J, KIM W Y, YUN D J. A new insight of salt stress signaling in plant[J]. Molecular Cells, 2016, 39(6): 447-459. DOI:10.14348/molcells.2016.0083 |
| [5] |
CHEESEMAN J M. The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions[J]. New Phytologist, 2015, 206(2): 557-570. DOI:10.1111/nph.13217 |
| [6] |
崔纪超, 中奕, 钟玉扬, 余金姜, 武小霞. 不同甘薯品种苗期耐盐性试验[J]. 广东农业科学, 2020, 47(4): 1-7. DOI:10.16768/j.issn.1004-874X.2020.04.001 CUI J C, ZHONG Y, ZHONG Y Y, YU J J, WU X X. Salt tolerance trial of different sweet potato varieties at seedling stage[J]. Guangdong Agricultural Sciences, 2020, 47(4): 1-7. DOI:10.16768/j.issn.1004-874X.2020.04.001 |
| [7] |
詹振楠, 王文娟. 种子引发对盐胁迫枸杞种子萌发的影响[J]. 广东农业科学, 2018, 45(6): 14-18. DOI:10.16768/j.issn.1004-874X.2018.06.003 ZHAN Z N, WANG W J. Effects of seed priming on Lycium barbarum seed germination under salt stress[J]. Guangdong Agricultural Sciences, 2018, 45(6): 14-18. DOI:10.16768/j.issn.1004-874X.2018.06.003 |
| [8] |
肖京林, 覃美, 凌桂芝, 黎晓峰. 植物细胞壁对有害金属与盐分耐受性作用研究进展[J]. 广东农业科学, 2020, 47(9): 73-80. DOI:10.16768/j.issn.1004-874X.2020.09.010 XIAO J L, QIN M, LING G Z, LI X F. Advances in studies on the resistance of plant cell walls to harmful metals and salt[J]. Guangdong Agricultural Sciences, 2020, 47(9): 73-80. DOI:10.16768/j.issn.1004-874X.2020.09.010 |
| [9] |
GE Z, DRESSELHAUS T, QU L J. How CrRLK1L receptor complexes perceive RALF signal[J]. Trends in Plant Science, 2019, 24(11): 978-981. DOI:10.1016/j.tplants.2019.09.002 |
| [10] |
HARUTA M, SABAT G, STECKER K, MINKOFF B B, SUSSMAN M R. A peptide hormone and its receptor protein kinase regulate plant cell expansion[J]. Science, 2014, 343(6169): 408-11. DOI:10.1126/science.1244454 |
| [11] |
STEGMANN M, MONAGHAN J, SMAKOWSKA-LUZAN E, ROVENICH H, LEHNER A, HOLTON N, BELKHADIR Y, ZIPFEL C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling[J]. Science, 2017, 355(6322): 287-289. DOI:10.1126/science.aal2541 |
| [12] |
ZHAO C, ZAYED O, YU Z, JIANG W, ZHU P, HSU C C, ZHANG L, TAO W A, LOZANO-DURAN R, ZHU J K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(51): 13123-13128. DOI:10.1073/pnas.1816991115 |
| [13] |
ZHOU H P, XIAO F, ZHENG Y, LIU G, ZHUANG Y, WNAG Z, ZHANG Y, HE J, FU C, LIN H H. PAMP-INDUCED SECRETED PEPTIDE 3 modulates salt tolerance through RECEPTOR-LIKE KINASE 7 in plants[J]. The Plant Cell, 2022, 34: 927-944. DOI:10.1093/plcell/koab292 |
| [14] |
ZHANG S S, SUN L, DONG X, LU S J, TIAN W, LIU J X. Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis[J]. Journal of Integrative Plant Biology, 2016, 58(7): 623-626. DOI:10.1111/jipb.12442 |
| [15] |
ENDLER A, KESTEN C, SCHNEIDER R, ZHANG Y, IVAKOV A, FROEHLICH A, FUNKE N, PERSSON S. A mechanism for sustained cellulose synthesis during salt stress[J]. Cell, 2015, 162(6): 1353-1364. DOI:10.1016/j.cell.2015.08.028 |
| [16] |
VAN DER DOES D, BOUTROT F, ENGELSDORF T, RHODES J, MCKENNA J F, VERNHETTES S, KOEVOETS I, TINTOR N, VEERABAGU M, MIEDES E, SEGONZAC C, ROUX M, BREDA A S, HARDTKE C S, MOLINA A, REP M, TESTERINK C, MOUILLE G, HOFTE H, HAMANN T, ZIPFEL C. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses[J]. PLoS Genetics, 2017, 13(6): e1006832. DOI:10.1371/journal.pgen.1006832 |
| [17] |
HEMATY K, SADO P E, VAN TUINEN A, ROCHANGE S, DESNOS T, BALZERGUE S, PELLETIER S, RENOU J P, HOFTE H. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis[J]. Current Biology, 2007, 17(11): 922-931. DOI:10.1016/j.cub.2007.05.018 |
| [18] |
YAN J W, LIU Y, YANG L, HE H, HUANG Y, FANG L, SCHELLER H V, JIANG M Y, ZHANG A Y. Cell wall β-1,4-galactan regulated by BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana[J]. Molecular Plant, 2021, 14(3): 411-425. DOI:10.1016/j.molp.2020.11.023 |
| [19] |
YAN J W, LIU Y, YAN J W, LIU Z H, LOU H Q, WU J S. The salt-activated CBF1/CBF2/CBF3-GALS1 module fine-tunes galactan-induced salt hypersensitivity in Arabidopsis[J]. Journal of Integrative Plant Biology, 2023, 65(8): 1904-1917. DOI:10.1111/jipb.13501 |
| [20] |
KIM H J, TRIPLETT B. Involvement of extracellular Cu/Zn superoxide dismutase in cotton fiber primary and secondarycell wall biosynthesis[J]. Plant Signaling Behavior, 2008, 3(12): 1119-1121. DOI:10.4161/psb.3.12.7039 |
| [21] |
LI Y X, ZHANG T T, KANG Y, WANG P, YU W, WANG J, LI W, JIANG X Y, ZHOU Y. Integrated metabolome, transcriptome analysis, and multi-f lux full-length sequencing offer novel insights into the function of lignin biosynthesis as a Sesuvium portulacastrum response to salt stress[J]. International Journal of Biological Macromolecules, 2023, 237: 124222. DOI:10.1016/j.ijbiomac.2023.124222 |
| [22] |
ZHAO C, ZAYED O, ZENG F, LIU C, ZHANG L, ZHU P, HSU C C, TUNCIL Y E, TAO W A, CARPITA N C, ZHU J K. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis[J]. New Phytologist, 2019, 224(1): 274-290. DOI:10.1111/nph.15867 |
| [23] |
DOBLAS V G, GELDNER N, BARBERON M. The endodermis, a tightly controlled barrier for nutrients[J]. Current Opinion in Plant Biology, 2017, 39: 136-143. DOI:10.1016/j.pbi.2017.06.010 |
| [24] |
VAN ZELM E, ZHANG Y, TESTERINK C. Salt tolerance mechanisms of plants[J]. Annual Review of Plant Biology, 2020, 71: 403-433. DOI:10.1146/annurev-arplant-050718-100005 |
| [25] |
BLUMWALD E, AHARON G S, APSE M P. Sodium transport in plant cells[J]. Biochim Biophys Acta, 2000, 1465(1-2): 140-151. DOI:10.1016/s0005-2736(00)00135-8 |
| [26] |
TUTEJA N. Mechanisms of high salinity tolerance in plants[J]. Methods in Enzymology, 2007, 428: 419-438. DOI:10.1016/S0076-6879(07)28024-3 |
| [27] |
YANG Y, GUO Y. Elucidating the molecular mechanisms mediating plant salt-stress responses[J]. New Phytologist, 2018, 217(2): 523-539. DOI:10.1111/nph.14920 |
| [28] |
JIANG Z H, ZHOU X, TAO M, YUAN F, LIU L, WU F, WU X, XIANG Y, NIU Y, LIU F, LI C, YE R, BYEON B, XUE Y, ZHAO H, WANG H N, CRAWFORD B M, JOHNSON D M, HU C, PEI C, ZHOU W, SWIFT G B, ZHANG H, VO-DINH T, HU Z, SIEDOW J N, PEI Z M. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx[J]. Nature, 2019, 572: 341-346. DOI:10.1038/s41586-019-1449-z |
| [29] |
ESSAH P A, DAVENPORT R, TESTER M. Sodium influx and accumulation in Arabidopsis[J]. Plant Physiology, 2003, 133(1): 307-318. DOI:10.1104/pp.103.022178 |
| [30] |
MUNNS R, TESTER M. Mechanisms of salinity tolerance[J]. Annual Review of Plant Biology, 2008, 59: 651-681. DOI:10.1146/annurev.arplant.59.032607.092911 |
| [31] |
FRANCISCO R, WALTER G, JULIAN I S. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance[J]. Science, 1995, 270(5242): 1660-1663. DOI:10.1126/science.270.5242.1660 |
| [32] |
RUS A, LEE B H, MUÑOZ-MAYOR A, SHARKHUU A, MIURA K, ZHU J K, BRESSAN R A, HASEGAWA P M. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta[J]. Plant Physiology, 2004, 136(1): 2500-2511. DOI:10.1104/pp.104.042234 |
| [33] |
RUS A, YOKOI S, SHARKHUU A, REDDY M, LEE B H, MATSUMOTO T K, KOIWA H, ZHU J K, BRESSAN R A, HASEGAWA P M. AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(24): 14150-14155. DOI:10.1073/pnas.241501798 |
| [34] |
UOZUMI N, KIM E J, RUBIO F, YAMAGUCHI T, MUTO S, TSUBOI A, BAKKER E P, NAKAMURA T, SCHROEDER J I. The Arabidopsis HKT1 gene homolog mediates inward Na(+) currents in xenopus laevis oocytes and Na(+) uptake in Saccharomyces cerevisiae[J]. Plant Physiology, 2000, 122(4): 1249-1259. DOI:10.1104/pp.122.4.1249 |
| [35] |
BERTHOMIEU P, CONÉJÉRO G, NUBLAT A, BRACKENBURY WJ, LAMBERT C, SAVIO C, UOZUMI N, OIKI S, YAMADA K, CELLIER F, GOSTI F, SIMONNEAU T, ESSAH PA, TESTER M, VÉRY AA, SENTENAC H, CASSE F. Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance[J]. The EMBO Journal, 2003, 22(9): 2004-2014. DOI:10.1093/emboj/cdg207 |
| [36] |
HAMAMOTO S, HORIE T, HAUSER F, DEINLEIN U, SCHROEDER J I, UOZUMI N. HKT transporters mediate salt stress resistance in plants: From structure and function to the field[J]. Current Opinion in Biotechnology, 2015, 32: 113-120. DOI:10.1016/j.copbio.2014.11.025 |
| [37] |
ALI A, MAGGIO A, BRESSAN R A, YUN D J. Role and functional differences of HKT1-type transporters in plants under salt stress[J]. International Journal of Molecular Sciences, 2019, 20(5): 1059. DOI:10.3390/ijms20051059 |
| [38] |
RUBIO F, GASSMANN W, SCHROEDER J I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance[J]. Science, 1995, 270(5242): 1660-1663. DOI:10.1126/science.270.5242.1660 |
| [39] |
SCHACHTMAN D P, SCHROEDER J I. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants[J]. Nature, 1994, 370(6491): 655-658. DOI:10.1038/370655a0 |
| [40] |
ZHANG J L, SHI H. Physiological and molecular mechanisms of plant salt tolerance[J]. Photosynthesis Research, 2013, 115(1): 1-22. DOI:10.1007/s11120-013-9813-6 |
| [41] |
KOBAYASHI N I, YAMAJI N, YAMAMOTO H, OKUBO K, UENO H, COSTA A, TANOI K, MATSUMURA H, FUJII-KASHINO M, HORIUCHI T, NAYEF M A, SHABALA S, AN G, MA J F, HORIE T. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice[J]. The Plant Journal, 2017, 91(4): 657-670. DOI:10.1111/tpj.13595 |
| [42] |
REN Z H, GAO J P, LI L G, CAI X L, HUANG W, CHAO D Y, ZHU M Z, WANG Z Y, LUAN S, LIN H X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter[J]. Nature Genetics, 2005, 37(10): 1141-1146. DOI:10.1038/ng1643 |
| [43] |
BYRT C S, PLATTEN J D, SPIELMEYER W, JAMES R A, LAGUDAH E S, DENNIS E S, TESTER M, MUNNS R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1[J]. Plant Physiology, 2007, 143(4): 1918-1928. DOI:10.1104/pp.106.093476 |
| [44] |
MUNNS R, JAMES RA, XU B, ATHMAN A, CONN SJ, JORDANS C, BYRT CS, HARE RA, TYERMAN SD, TESTER M, PLETT D, GILLIHAM M. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene[J]. Nature Biotechnology, 2012, 30(4): 360-364. DOI:10.1038/nbt.2120 |
| [45] |
杨玲琴, 刘敬, 李魏, 戴良英. 植物钾离子通道AKT1的研究进展[J]. 生物技术通报, 2019, 35(4): 94-100. DOI:10.13560/j.cnki.biotech.bull.1985.2019-0033 YANG L Q, LIU J, LI W, DAI L Y. Research advances in potassium ion channel AKT1 in plant[J]. Biotechnology Bulletin, 2019, 35(4): 94-100. DOI:10.13560/j.cnki.biotech.bull.1985.2019-0033 |
| [46] |
NIEVES-CORDONES M, MILLER A J, ALEMAN F, MARTINEZ V, RUBIO F. A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5[J]. Plant Molecular Biology, 2008, 68(6): 521-532. DOI:10.1007/s11103-008-9388-3 |
| [47] |
GOLLDACK D, QUIGLEY F, MICHALOWSKI C B, KAMASANI U R, BOHNERT H J. Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently[J]. Plant Molecular Biology, 2003, 51: 71-81. DOI:10.1023/A:1020763218045 |
| [48] |
DUAN H R, MA Q, ZHANG J L, HU J, BAO A K, WEI L, WANG Q, LUAN S, WANG S M. The inward-rectifying K+ channel SsAKT1 is a candidate involved in K+ uptake in the halophyte Suaeda salsa under saline condition[J]. Plant Soil, 2015, 395: 173-187. DOI:10.1007/s11104-015-2539-9 |
| [49] |
ARDIE S W, LIU S, TAKANO T. Expression of the AKT1-type K(+) channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis[J]. Plant Cell Reports, 2010, 29(8): 865-874. DOI:10.1007/s00299-010-0872-2 |
| [50] |
ZEESHAN M, LU M, NAZ S, SEHAR S, CAO F, WU F. Resemblance and difference of seedling metabolic and transporter gene expression in high tolerance wheat and barley cultivars in response to salinity stress[J]. Plants (Basel), 2020, 9(4): 519. DOI:10.3390/plants9040519 |
| [51] |
JOSHI S, NATH J, SINGH A K, PAREEK A, JOSHI R. Ion transporters and their regulatory signal transduction mechanisms for salinity tolerance in plants[J]. Physiologia Plantarum, 2022, 174: e13702. DOI:10.1111/ppl.13702 |
| [52] |
BASU S, KUMAR A, BENAZIR I, KUMAR G. Reassessing the role of ion homeostasis for improving salinity tolerance in crop plants[J]. Physiologia Plantarum, 2021, 171(4): 502-519. DOI:10.1111/ppl.13112 |
| [53] |
KRONZUCKER H J, BRITTO D T. Sodium transport in plants: A critical revies[J]. New Phytologist, 2011, 189: 54-81. DOI:10.1111/j.1469-8137.2010.03540.x |
| [54] |
MAATHUIS F J, SANDERS D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides[J]. Plant Physiology, 2001, 127(4): 1617-1625. DOI:10.1104/pp.010502 |
| [55] |
WU S J, DING L, ZHU J K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition[J]. The Plant Cell, 1996, 8(4): 617-627. DOI:10.1105/tpc.8.4.617 |
| [56] |
ZHU J K, LIU J, XIONG L. Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition[J]. The Plant Cell, 1998, 10(7): 1181-1191. DOI:10.1105/tpc.10.7.1181 |
| [57] |
QIU Q S, GUO Y, DIETRICH M A, SCHUMAKER K S, ZHU J K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(12): 8436-8441. DOI:10.1073/pnas.122224699 |
| [58] |
SHI H, QUINTERO F J, PARDO J M, ZHU J K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants[J]. The Plant Cell, 2002, 14: 465-477. DOI:10.1105/tpc.010371 |
| [59] |
QUINTERO F J, MARTINEZ-ATIENZA J, VILLALTA I, JIANG X, KIM W Y, ALI Z, FUJII H, MENDOZA I, YUN D J, ZHU J K, PARDO J M. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(6): 2611-2616. DOI:10.1073/pnas.1018921108 |
| [60] |
SHI H, ISHITANI M, KIM C, ZHU J K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97: 6896-6901. DOI:10.1073/pnas.120170197 |
| [61] |
MARTINEZ-ATIENZA J, JIANG X, GARCIADEBLAS B, MENDOZA I, ZHU J K, PARDO J M, QUINTERO F J. Conservation of the salt overly sensitive pathway in rice[J]. Plant Physiology, 2007, 143(2): 1001-1012. DOI:10.1104/pp.106.092635: |
| [62] |
XU H, JIANG X Y, ZHAN K H, CHENG X Y, CHEN X J, PARDO J M, CUI D. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast[J]. Archives of Biochemistry and Biophysics, 2008, 473(1): 8-15. DOI:10.1016/j.abb.2008.02.018 |
| [63] |
YIN X C, XIA Y Q, XIE Q, CAO Y X, WANG Z Y, HAO G P, SONG J, ZHOU Y, JIANG X Y. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance[J]. Journal of Experimental Botany, 2020, 71(6): 1801-1814. DOI:10.1093/jxb/erz549 |
| [64] |
TANG R J, LIU H, BAO Y, LV Q D, YANG L, ZHANG H X. The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress[J]. Plant Molecular Biology, 2010, 74(4-5): 367-380. DOI:10.1007/s11103-010-9680-x |
| [65] |
OH D H, LEIDI E, ZHANG Q, HWANG S M, LI Y, QUINTERO F J, JIANG X, D'URZO M P, LEE S Y, ZHAO Y, BAHK J D, BRESSAN R A, YUN D J, PARDO J M, BOHNERT H J. Loss of halophytism by interference with SOS1 expression[J]. Plant Physiology, 2009, 151(1): 210-22. DOI:10.1104/pp.109.137802 |
| [66] |
ZHOU Y, ZHU Y, LI W, ZHANG T T, LI Y X, KANG Y Q, WANG J, GUO J C, JIANG X Y. Heterologous expression of Sesuvium portulacastrum SOS-related genes confer salt tolerance in yeast[J]. Acta Physiologiae Plantarum, 2023, 45(4): 58. DOI:10.1007/s11738-023-03518-7 |
| [67] |
YANG Z, WANG C, XUE Y, LIU X, CHEN S, SONG C P, YANG Y Q, GUO Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance[J]. Nature Communication, 2019, 10: 1199. DOI:10.1038/s41467-019-09181-2 |
| [68] |
MA L, YE J M, YANG Y Q, LIN H X, YUE L L, LUO J, LONG Y, FU H Q, LIU X N, ZHANG Y L, WANG Y, CHEN L Y, KUDLA J, WANG Y J, HAN S C, SONG C P, GUO Y. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress[J]. Development Cell, 2019, 48: 697-709. DOI:10.1016/j.devcel.2019.02.010 |
| [69] |
LI Q, FU H, YU X, WEN X, GUO H W, GUO Y, LI J R. The SALT OVERLY SENSITIVE 2-CONSTITUTIVE TRIPLE RESPONSE1 module coordinates plant growth and salt tolerance in Arabidopsis[J]. Journal of Experimental Botany, 2023, erad368. DOI:10.1093/jxb/erad368 |
| [70] |
XIE Q, ZHOU Y, JIANG X Y. Structure, function, and regulation of the plasma membrane Na+/H+ antiporter salt overly sensitive 1 in plants[J]. Frontiers in Plant Science, 2022, 13: 866265. DOI:10.3389/fpls.2022.866265 |
| [71] |
WANG Y, PAN C, CHEN Q, XIE Q, GAO Y, HE L, LI Y, DONG Y, JIANG X Y, ZHAO Y. Architecture and autoinhibitory mechanism of the plasma membrane Na+/H+ antiporter SOS1 in Arabidopsis[J]. Nature Communication, 2023, 14(1): 4487. DOI:10.1038/s41467-023-40215-y |
| [72] |
QUINTERO F J, OHTA M, SHI H, ZHU J K, PARDO J M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(13): 9061-6. DOI:10.1073/pnas.132092099 |
| [73] |
NÚÑEZ-RAMÍREZ R, SÁNCHEZ-BARRENA M J, VILLALTA I, VEGA J F, PARDO J M, QUINTERO F J, MARTINEZ-SALAZAR J, ALBERT A. Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na(+)/H(+) antiporter[J]. Journal of Molecular Biology, 2012, 424(5): 283-294. DOI:10.1016/j.jmb.2012.09.015 |
| [74] |
JARVIS D E, RYU C H, BEILSTEIN M A, SCHUMAKER K S. Distinct roles for SOS1 in the convergent evolution of salt tolerance in Eutrema salsugineum and Schrenkiella parvula[J]. Molecular Biology and Evolution, 2014, 31: 2094-2107. DOI:10.1093/molbev/msu152 |
| [75] |
GUO Y, HALFTER U, ISHITANI M, ZHU J K. Molecula r characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance[J]. The Plant Cell, 2001, 13: 1383-1400. DOI:10.1105/TPC.010021 |
| [76] |
GONG D, GUO Y, JAGENDORF A T, ZHU J K. Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance[J]. Plant Physiology, 2002, 130(1): 256-264. DOI:10.1104/pp.004507 |
| [77] |
ZHOU Y, LAI Z, YIN X, YU S, XU Y, WANG X, CONG X, LUO Y, XU H X, JIANG X Y. Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco[J]. Plant Science, 2016, 253: 176-186. DOI:10.1016/j.plantsci.2016.09.016 |
| [78] |
马瑞, 李世贵, 刘维刚, 王志科, 杨江伟, 唐勋, 张宁, 司怀军. 植物CBL-CIPK信号系统的功能及其响应非生物胁迫作用机制研究进展[J]. 植物生理学报, 2021, 57(3): 521-530. DOI:10.13592/j.cnki.ppj.2020.0173 MA R, LI S G, LIU W G, WANG Z K, YANG J W, TANG X, ZHANG N, SI H J. Functions and progress in mechanism research of CBL-CIPK signaling system in plants[J]. Plant Physiology Journal, 2021, 57(3): 521-530. DOI:10.13592/j.cnki.ppj.2020.0173 |
| [79] |
TANG R J, YANG Y, YANG L, LIU H, WANG C T, YU M M, GAO X S, ZHANG H X. Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane[J]. Plant Cell and Environment, 2014, 37(3): 573-588. DOI:10.1111/pce.12178 |
| [80] |
HU D G, LI M, LUO H, DONG Q L, YAO Y X, YOU C X, HAO Y J. Molecular cloning and functional characterization of MdSOS2 reveals its involvement in salt tolerance in apple callus and Arabidopsis[J]. Plant Cell Reports, 2012, 31(4): 713-722. DOI:10.1007/s00299-011-1189-5 |
| [81] |
HUERTAS R, OLÍAS R, ELJAKAOUI Z, GÁLVEZ F J, LI J, DE MORALES P A, BELVER A, RODRÍGUEZ-ROSALES M P. Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato[J]. Plant Cell and Environment, 2012, 35(8): 1467-1482. DOI:10.1111/j.1365-3040.2012.02504.x |
| [82] |
QUAN R, LIN H, MENDOZA I, ZHANG Y, CAO W, YANG Y, SHANG M, CHEN S, PARDO J M, GUO Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress[J]. The Plant Cell, 2007, 19(4): 1415-1431. DOI:10.1105/tpc.106.042291 |
| [83] |
KIM B G, WAADT R, CHEONG Y H, PANDEY G K, DOMINGUEZ-SOLIS J R, SCHÜLTKE S, LEE S C, KUDLA J, LUAN S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis[J]. The Plant Journal, 2007, 52(3): 473-484. DOI:10.1111/j.1365-313X.2007.03249.x |
| [84] |
BLUMWALD E, POOLE R J. Na+/H+ antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris[J]. Plant Physiology, 1985, 78: 163-167. DOI:10.1104/pp.78.1.163 |
| [85] |
GONG Z Z, XIONG L M, SHI H Z, YANG S H, HERRERA-ESTRELLA L R, XU G H, CHAO D Y, LI J R, WANG P Y, QIN F, LI J J, DING Y L, SHI Y T, WANG Y, YANG Y Q, GUO Y, ZHU J K. Plant abiotic stress response and nutrient use efficiency[J]. Science China Life Sciences, 2020, 63(5): 635-674. DOI:10.1007/s11427-020-1683-x |
| [86] |
GAXIOLA R A, RAO R, SHERMAN A, GRISAFI P, ALPER S L, FINK G R. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast[J]. Proceedings of The National Academy of Sciences of The United States of America, 1999, 96(4): 1480-1485. DOI:10.1073/pnas.96.4.1480 |
| [87] |
APSE M P, AHARON G S, SNEDDEN W A, BLUMWALD E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis[J]. Science, 1999, 285(5431): 1256-1258. DOI:10.1126/science.285.5431.1256 |
| [88] |
ZHANG H X, BLUMWALD E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit[J]. Nature Biotechnology, 2001, 19(8): 765-768. DOI:10.1038/90824 |
| [89] |
ZHANG H X, HODSON J N, WILLIAMS J P, BLUMWALD E. Engineering salt-tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation[J]. Proceedings of The National Academy of Sciences of The United States of America, 2001, 98(22): 12832-12836. DOI:10.1073/pnas.231476498 |
| [90] |
YOKOI S, QUINTERO F J, CUBERO B, RUIZ M T, BRESSAN R A, HASEGAWA P M, PARDO J M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response[J]. The Plant Journal, 2002, 30(5): 529-539. DOI:10.1046/j.1365-313x.2002.01309.x |
| [91] |
BASSIL E, TAJIMA H, LIANG Y C, OHTO M A, USHIJIMA K, NAKANO R, ESUMI T, COKU A, BELMONTE M, BLUMWALD E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction[J]. The Plant Cell, 2011, 23(9): 3482-3497. DOI:10.1105/tpc.111.089581 |
| [92] |
BASSIL E, OHTO M A, ESUMI T, TAJIMA H, ZHU Z, CAGNAC O, BELMONTE M, PELEG Z, YAMAGUCHI T, BLUMWALD E. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development[J]. The Plant Cell, 2011, 23(1): 224-239. DOI:10.1105/tpc.110.079426 |
| [93] |
AN R, CHEN Q J, CHAI M F, LU P L, SU Z, QIN Z X, CHEN J, WANG X C. ATNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter[J]. The Plant Journal, 2007, 49(4): 718-728. DOI:10.1111/j.1365-313X.2006.02990.x |
| [94] |
YAMAGUCHI T, AHARON G S, SOTTOSANTO J B, BLUMWALD E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(44): 16107-16112. DOI:10.1073/pnas.0504437102 |
| [95] |
YAMAGUCHI T, APSE MP, SHI H, BLUMWALD E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(21): 12510-12515. DOI:10.1073/pnas.2034966100 |
| [96] |
PABUAYON I C M, JIANG J, QIAN H, CHUNG J S, SHI H Z. Gain-of-function mutations of AtNHX1 suppress sos1 salt sensitivity and improve salt tolerance in Arabidopsis[J]. Stress Biology, 2021, 1: 14. DOI:10.1007/s44154-021-00014-1 |
| [97] |
BAO A K, WANG S M, WU G Q, XI J J, ZHANG J L, WANG C M. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.)[J]. Plant Science, 2009, 176: 232-240. DOI:10.1016/J.PLANTSCI.2008.10.009 |
| [98] |
LIU S P, ZHENG L Q, XUE Y H, ZHANG Q, WANG L, SHOU H X. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice[J]. Journal of Plant Biology, 2010, 53(6): 444-452. DOI:10.1007/s12374-010-9135-6 |
| [99] |
ZHAO F Y, ZHANG X J, LI P H, ZHAO Y X, ZHANG H. Co-expression of the Suaeda salsa SsNHX1 and Arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single SsNHX1[J]. Molecular Breeding, 2006, 17(4): 341-353. DOI:10.1007/s11032-006-9005-6 |
| [100] |
BRINI F, HANIN M, MEZGHANI I, BERKOWITZ G A, MASMOUDI K. Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants[J]. Journal of Experimental Botany, 2007, 58(2): 301-308. DOI:10.1093/jxb/erl251 |
| [101] |
GOUIAA S, KHOUDI H, LEIDI EO, PARDO J M, MASMOUDI K. Expression of wheat Na+/H+ antiporter TNHXS1 and H+-pyrophosphatase TVP1 genes in tobacco from a bicistronic transcriptional unit improves salt tolerance[J]. Plant Molecular Biology Reporter, 2012, 79(1-2): 137-155. DOI:10.1007/s11103-012-9901-6 |
| [102] |
ZHOU Y G, XU K, GAO H, YAO W, ZHANG Y, ZHANG Y, HUSSAIN M A, WANG F, YANG X, LI H Y. Comparative proteomic analysis of two wild soybean (Glycine soja) genotypes reveals positive regulation of saline-alkaline stress tolerance by tonoplast transporters[J]. Journal of Agricultural and Food Chemistry, 2023, 71: 14109-14124. DOI:10.1021/acs.jafc.3c02111 |
| [103] |
QIU Q S, GUO Y, QUINTERO F J, PARDO J M, SCHUMAKER K S, ZHU J K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway[J]. Journal of Biological Chemistry, 2004, 279: 207-215. DOI:10.1074/jbc.M307982200 |
| [104] |
BATELLI G, VERSLUES P E, AGIUS F, QIU Q, FUJII H, PAN S, SCHUMAKER K S, GRILLO S, ZHU J K. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity[J]. Molecular and Cellular Biology, 2007, 27(22): 7781-7790. DOI:10.1128/MCB.00430-07 |
| [105] |
KIM B G, WAADT R, CHEONG Y H, PANDEY G K, DOMINGUEZ-SOLIS J R, SCHÜLTKE S, LEE S C, KUDLA J, LUAN S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis[J]. The Plant Journal, 2007, 52(3): 473-484. DOI:10.1111/j.1365-313X.2007.03249.x |
| [106] |
PANDEY G K, KANWAR P, SINGH A, STEINHORST L, LUAN S. Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis[J]. Plant Physiology, 2015, 169: 780-792. DOI:10.1104/pp.15.00623 |
| [107] |
KHAN S A, LI M Z, WANG S M, YIN H J. Revisiting the role of plant transcription factors in the battle against abiotic stress[J]. International Journal of Molecular Sciences, 2018, 19(6): 1634. DOI:10.3390/ijms19061634 |
| [108] |
KREPS J A, WU Y, CHANG H S, ZHU T, WANG X, HARPER J F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress[J]. Plant Physiology, 2002, 130: 1-21. DOI:10.1104/pp.008532 |
| [109] |
ZELLER G, HENZ S R, WIDMER C K, SACHSENBERG T, RÄTSCH G, WEIGEL D, LAUBINGER S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole genome tiling arrays[J]. The Plant Joural, 2009, 58: 1068-1082. |
| [110] |
KIM J H, NGUYEN N H, JEONG C Y, NGUYEN N T, HONG S W, LEE H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis[J]. Journal of Plant Physiology, 2013, 170(16): 1461-1465. DOI:10.1016/j.jplph.2013.05.011 |
| [111] |
DOSSA K, MMADI M A, ZHOU R, LIU A, YANG Y, DIOUF D, YOU J, ZHANG X. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis[J]. AoB Plants, 2019, 12(1): plz081. DOI:10.1093/aobpla/plz081 |
| [112] |
CHEN H, LAI L, LI L, LIU L, JAKADA B H, HUANG Y, HE Q, CHAI M, NIU X, QIN Y. AcoMYB4, an Ananas comosus L. MYB transcription factor, functions in osmotic stress through negative regulation of ABA signaling[J]. International Journal of Molecular Sciences, 2020, 21(16): 5727. DOI:10.3390/ijms21165727 |
| [113] |
FANG Q, JIANG T, XU L, LIU H, MAO H, WANG X, JIAO B, DUAN Y, WANG Q, DONG Q, YANG L, TIAN G, ZHANG C, ZHOU Y, LIU X, WANG H, FAN D, WANG B, LUO K. A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis[J]. Plant Physiology Biochemistry, 2017, 114: 100-110. DOI:10.1016/j.plaphy.2017.02.018 |
| [114] |
SONG Y, LI J, SUI Y, HAN G, ZHANG Y, GUO S, SUI N. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis[J]. Plant Molecular Biology, 2020, 102(6): 603-614. DOI:10.1007/s11103-020-00966-4 |
| [115] |
DAI W, WANG M, GONG X, LIU J H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs[J]. New Phytologist, 2018, 219(3): 972-989. DOI:10.1111/nph.15240 |
| [116] |
AN J P, YAO J F, XU R R, YOU C X, WANG X F, HAO Y J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response[J]. Physiologia Plantarum, 2018, 164(3): 279-289. DOI:10.1111/ppl.12724 |
| [117] |
YANG X, KIM M Y, HA J, LEE S H. Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants[J]. Frontiers in Plant Science, 2019, 10: 1036. DOI:10.3389/fpls.2019.01036 |
| [118] |
WANG Q, GUO C, LI Z, SUN J, DENG Z, WEN L, LI X, GUO Y. Potato NAC transcription factor StNAC053 enhances salt and drought tolerance in transgenic Arabidopsis[J]. International Journal of Molecular Sciences, 2021, 22(5): 2568. DOI:10.3390/ijms22052568 |
| [119] |
LI Q, WU Q, WANG A, LV B, DONG Q, YAO Y, WU Q, ZHAO H, LI C, CHEN H, WANG X. Tartary buckwheat transcription factor FtbZIP83 improves the drought/salt tolerance of Arabidopsis via an ABA-mediated pathway[J]. Plant Physiology Biochemistry, 2019, 144: 312-323. DOI:10.1016/j.plaphy.2019.10.003 |
| [120] |
LI Q, ZHAO H, WANG X, KANG J, LV B, DONG Q, LI C, CHEN H, WU Q. Tartary buckwheat transcription factor FtbZIP5, regulated by FtSnRK2.6, can improve salt/drought resistance in transgenic Arabidopsis[J]. International Journal of Molecular Sciences, 2020, 21(3): 1123. DOI:10.3390/ijms21031123 |
| [121] |
MA L, HAN R, YANG Y Q, LIU X, LI H, ZHAO X, LI J, FU H, HUO Y, SUN L, YAN Y, ZHANG H Y, LI Z, TIAN F, LI J L, GUO Y. Phytochromes enhance SOS2-mediated PIF1 and PIF3 phosphorylation and degradation to promote Arabidopsis salt tolerance[J]. The Plant Cell, 2023, 35(8): 2997-3020. DOI:10.1093/plcell/koad117 |
| [122] |
YANG R, YANG Z, XING M, JING Y, ZHANG Y, ZHANG K, ZHOU Y, ZHAO H, QIAO W, SUN J. TaBZR1 enhances wheat salt tolerance via promoting ABA biosynthesis and ROS scavenging[J]. Journal of Genetics and Genomics, 2023. DOI:10.1016/j.jgg.2023.09.006 |
| [123] |
LUAN S, KUDLA J, RODRIGUEZ-CONCEPCION M, YALOVSKY S, GRUISSEM W. Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants[J]. The Plant Cell, 2002, 14: S389-S400. DOI:10.1105/tpc.001115 |
| [124] |
WU G Q, FENG R J, WANG S M, WANG C M, BAO A K, WEI L, YUAN H J. Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 confers enhanced salinity tolerance in chimeric sugar beet (Beta vulgaris L.)[J]. Frontiers in Plant Science, 2015, 6: 581. DOI:10.3389/fpls.2015.00581 |
| [125] |
MA D M, WR W X, LI H W, JIN F X, GUO L N, WANG J, DA H J, XU X. Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.)[J]. Protoplasma, 2014, 251(1): 219-231. DOI:10.1007/s00709-013-0540-9 |
| [126] |
ZHOU Y, YIN X, DUAN R, GUO J, JIANG X Y. SpAHA1 and SpSOS1 coordinate in transgenic yeast to improve salt tolerance[J]. PLoS One, 2015, 10: e0137447. DOI:10.1371/journal.pone.0137447 |
| [127] |
FAN Y, YIN X, XIE Q, XIA Y, WANG Z, SONG J, ZHOU Y, JIANG X Y. Co-expression of SpSOS1 and SpAHA1 in transgenic Arabidopsis plants improves salinity tolerance[J]. BMC Plant Biology, 2019, 19(1): 74. DOI:10.1186/s12870-019-1680-7 |
| [128] |
FEKI K, QUINTERO F J, KHOUDI H, LEIDI E O, MASMOUDI K, PARDO J M, BRINI F. A constitutively active form of a durum wheat Na(+)/H(+) antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis[J]. Plant Cell Reports, 2014, 33(2): 277-288. DOI:10.1007/s00299-013-1528-9 |
| [129] |
CHENG C, ZHONG Y, WANG Q, CAI Z, WANG D, LI C. Genome-wide identification and gene expression analysis of SOS family genes in tuber mustard (Brassica juncea var. tumida)[J]. PLoS ONE, 2019, 14(11): e0224672. DOI:10.1371/journal.pone.0224672: |
| [130] |
NUTAN K K, KUMAR G, SINGLA-PAREEK S L, PAREEK A. A salt overly sensitive pathway member from Brassica juncea BjSOS3 can functionally complement ΔAtsos3 in Arabidopsis[J]. Current Genomics, 2018, 19(1): 60-69. DOI:10.2174/1389202918666170228133621 |
(责任编辑 马春敏)
 2023, Vol. 50
2023, Vol. 50