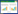文章信息
基金项目
- 河南省自然科学基金面上项目(232300420027);河南省人力资源和社会保障厅留学人员科研择优资助项目(2023039);河南省大学生创新项目(S202110480050)
作者简介
- 张晓东(1980—),男,博士,副教授,研究方向为道地药材次生代谢与调控,E-mail:zxd95@126.com.
文章历史
- 收稿日期:2023-01-23
【研究意义】禹白芷(Angelica dahurica cv. Yubaizhi)是河南道地药材,为伞形科当归属植物白芷的干燥根。禹白芷富含香豆素类、挥发油类和黄酮类化合物,具有解表散寒,祛风止痛,宣通鼻窍,燥湿止带,消肿排脓等功效,常被用于治疗感冒头痛、眉棱骨痛、鼻塞流涕、鼻鼽、鼻渊、牙痛、带下、疮疡肿痛等病症[1]。禹白芷位于禹药之首,被收录于历届《中国药典》[1]。根据《地理标志产品保护规定》,我国自2006年12月28日起对禹白芷实施地理标志产品保护。由于其独特的香气和优质的特性,禹白芷被广泛用于中医临床饮片配方、保健品、化妆品和香料等领域[2]。然而,禹白芷生产面临连作障碍和道地产区耕地面积逐年减少的问题。因此,利用合成生物学方法生产禹白芷的主要药效成分香豆素具有重要的前景。【前人研究进展】在植物中,香豆素的生物合成起源于苯丙素途径[3]。苯丙氨酸解氨酶(Phenylalanine ammonia-lyase,PAL,EC:4.3.1.24)是苯丙素生物合成途径的第一个酶(图 1),能够催化L- 苯丙氨酸中氨的氧化,生成反式肉桂酸,并释放氨气[4]。在植物中,PAL由其多基因家族编码[5]。如拟南芥有4个PAL基因[6]、石防风属植物(Kitagawia praeruptora)有3个PAL基因[7-8]、不同品种咖啡中发现3~6个PAL基因[9]、生菜有5个PAL基因[10]、马铃薯基因组中发现多达14个PAL基因[11],在禹白芷转录组中预测有4个PAL基因。目前,PAL基因已从拟南芥[12]、孜然芹[13]、罗勒[5]等许多植物中分离。PAL基因的表达具有组织特异性,并被生物和非生物因素诱导。例如,在前胡中,PpPAL基因主要在根中表达,茎和叶中也有少量表达,且茉莉酸甲酯、紫外线和冷处理均能够上调该基因的表达,而过氧化氢、热处理则下调该基因的表达[7];抽薹后,叶中PpPAL1基因表达量急剧上升,而叶中PpPAL2基因表达量急剧下降[8]。在咖啡中,叶锈病真菌可诱导PAL基因表达,与易感品种Caturra相比,抗性品种Híbrido de Timor的诱导率更高[9]。在孜然芹中,CcPAL在芽中表达量最高,其次是种子,根中的表达量最低,且镉、受伤、水杨酸和冷处理均可促进其表达[13]。生产上,PAL基因具有广泛用途。研究发现,PAL基因是鉴别金针菇颜色的关键基因[14]。改造后的拟南芥PAL可用于光学纯L- 苯丙氨酸的大规模生产[12]。喷洒壳聚糖纳米颗粒在水稻中可以提升PAL酶活性、降低水稻种子中苯丙氨酸的含量,为苯丙酮尿症患者提供低苯丙氨酸的稻米[15]。【本研究切入点】目前,禹白芷的研究主要集中在化学成分鉴定[16]、含量测定[17]以及主要药效成分的药理作用[18]等方面。然而,禹白芷AdPAL1基因尚未有相关报道。为实现在工程菌中高效生产禹白芷中的高价值香豆素成分,必须对香豆素生物合成途径的基因进行克隆和功能解析。【拟解决的关键问题】本研究拟首先测定禹白芷快速生长期和收获期根和叶中主要香豆素成分欧前胡素的含量,并同时测定AdPAL1基因同期的表达情况,探究它们之间的相关性。随后,根据禹白芷转录组中AdPAL1基因序列,设计特异性引物,利用RT-PCR技术从禹白芷根中扩增AdPAL1基因,并进行序列分析和原核表达,以期为禹白芷AdPAL1基因的功能及其在欧前胡素生物合成中的作用机制解析提供参考。
|
| 图 1 植物香豆素生物合成途径 Fig. 1 Coumarin biosynthesis pathway in plants |
1 材料与方法 1.1 试验材料
供试材料为禹州市中药材标准化中心提供的禹白芷(Angelica dahurica cv. Yubaizhi)种子。2020年9月5日,在许昌学院道地药材种植基地播种;并于2021年6月29日(快速生长期)和2021年7月29日(收获期)采样。基因克隆所用材料为禹白芷的根,基因组织特异性表达分析所用材料为一年生禹白芷的根(顶端1/5处)和成熟的大叶片。
1.2 试验方法1.2.1 欧前胡素含量测定 采用安捷伦1260高效液相色谱(HPLC)仪,按照《中国药典》[1]的方法测定白芷干燥根和干燥叶中欧前胡素的含量。使用梯度浓度绘制标准品的标准曲线,所有样品均重复3次。
1.2.2 根总RNA提取及AdPAL1基因开放阅读框(ORF)克隆 按照张玲等[19]的方法提取禹白芷根总RNA、合成cDNA和克隆AdPAL1基因ORF,获得重组质粒pMD19- AdPAL1。AdPAL1基因引物为AdPAL1NdeI-F:CATATGCTTGTGAGGATCAACACACT(下划线为NdeI酶切位点),AdPAL1HindIII-R:AAGCTTAGAAATTGGTAGAGGAGCTCCAT(下划线为HindIII酶切位点),退火温度为61 ℃。
1.2.3 AdPAL1基因原核表达载体构建 通过酶切连接的方法[19]构建原核表达载体ProS2-AdPAL1。
1.2.4 AdPAL蛋白生物信息学分析 按照张玲等[19]的方法对AdPAL1蛋白进行生物信息学分析。利用ProtComp v9.0软件预测蛋白的亚细胞定位情况。使用STRING网站对AdPAL1蛋白与其他蛋白间的互作进行预测[20]。多序列比对和进化树构建所使用蛋白的GenBank登录号分别为:绿竹Bambusa oldhamii,BoPAL(AAR24505.1);水稻Oryza sativa indica group,OsPAL(CAA61198.1);青稞Hordeum vulgare subsp. vulgare,HvPAL(CAA89007.1);小麦Triticum aestivum,TaPAL(CAA68036.1);小立碗藓Physcomitrella patens,PpaPAL(XP_001760495.1);银杏Ginkgo biloba,GbiPAL(ABU49842.1);麻黄Ephedra sinica,EsPAL(BAG74770.1);欧洲云杉Picea abies,PaPAL(CAK 22402.1);火炬松Pinus taeda,PtaPAL(AAA84889.1);罂粟Papaver somniferum,PsPAL(XP_026403210.1);毛果杨Populus trichocarpa,PtPAL(ACC63889.1);狭叶油茶Camellia lanceoleosa,ClPAL(KAI8012953.1);小叶种茶Camellia sinensis var. sinensis,CsPALd(QIH97409.1);石防风属植物Kitagawia praeruptora,KpPAL1(UXG20228.1)、KpPAL2(UXG20229.1);美味猕猴桃Actinidia deliciosa,AdePAL2(QED11031.1);中华猕猴桃Actinidia chinensis var. chinensis,AcPAL(PSS23706.1);紫花前胡Angelica decursiva,AdecPAL(QCY50324.1);野胡萝卜Daucus carota,DcPAL(BAC56977.1);欧芹Petroselinum crispum,PcPAL3(P45729.1);当归Angelica sinensis,AsPAL(AJW77399.1);金银花Lonicera japonica,LjPAL(AGE10589.1);灰毡毛忍冬Lonicera macranthoides,LmPAL(AZR37753.1);蓝靛忍冬Lonicera caerulea,LcPA(ALU09327.1);莴苣Lactuca sativa,LsPAL(XP_023767814.1);雪莲Saussurea involucrata,SiPAL(ALK02780.1);刺苞菜蓟Cynara cardunculus var. scolymus,CcPAL(XP_024986252.1);青蒿Artemisia annua,AaPAL(AKP55356.1);红凤菜Gynura bicolor,GbPAL(BAJ17655.1);西洋梨Pyrus communis,PcPAL(AGL81344.1);芝麻Sesamum indicum,SinPAL(XP_011077338.1);向日葵Helianthus annuus,HaPAL(KAJ0657456.1);非洲菊Gerbera jamesonii,GjPAL(QBC35985.1);苹果Malus domestica,MdPAL1(XP_008387584.2);紫苏Perilla frutescens,PfPAL(AEZ67457.1);禹白芷Angelica dahurica cv. Yubaizhi,AdAPL1(WGD08245.1),AdPAL2(WGD08246.1)。
1.2.5 AdPAL1基因的原核表达 参照张玲等[19]的方法进行重组质粒ProS2-AdPAL1的原核表达与SDS-PAGE检测。菌体收集后,使用1×PBS洗涤菌体,使用JY92-IIDN型超声细胞破碎仪(新芝,宁波)进行细胞破碎。4 ℃、12 000 r/min离心30 min,分离上清和沉淀。
1.2.6 AdPAL1基因的组织特异性表达分析 分别取一年生快速生长期和收获期禹白芷的根和叶,按照张玲等[19]的方法进行AdPAL1基因的组织特异性表达分析。选择AdSAND基因[21]作为内参,引物为qAdSAND-F:5'- TGCTGGAGGAAAGGGGAC-3' 和qAdSAND-R:5'- GCCATTTCCCAGGGACCC-3',PCR条件为:95 ℃ 30 s,95 ℃ 5 s,60 ℃ 20 s。AdPAL1引物为qAdPAL1-F:5'-AGTGCTAGGGCTGTGCTTG-3' 和qAdPAL1-R:5'-TCCTCTCCAGGCGATGTCA-3',PCR条件为:95℃ 30 s,95℃ 5 s,60℃ 20 s。每个反应重复3次。反应在Archimed X4荧光定量PCR仪(鲲鹏基因,北京)上进行扩增,扩增曲线、溶解曲线、标准曲线由定量PCR仪软件自动生成。使用内参基因校准后,分别计算根和叶中AdPAL1基因相对表达量。采用2-△△Ct的方法进行数据分析。
1.2.7 数据分析 使用Origin 2018C对欧前胡素含量和AdPAL1基因的表达量进行显著性分析。使用Excel 2019中的CORREL函数计算快速生长期与收获期欧前胡素变化量、AdPAL1基因表达变化量的相关系数。
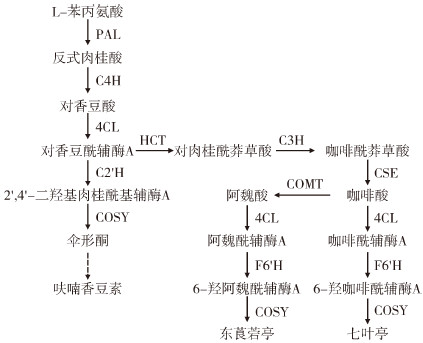
2 结果与分析 2.1 欧前胡素含量测定分别对禹白芷快速生长期和收获期根和叶中的欧前胡素含量进行测定,结果(图 2)表明,两个时期根中的欧前胡素含量均显著高于叶,并且在快速生长期根和叶中的欧前胡素含量均略高于收获期。
|
| **、*** 分别表示0.01、0.001水平上差异显著;NS表示差异不显著 **, *** indicate significant differences at 0.01, 0.001 levels, respectively; NS indicates that the difference is not significant 图 2 不同生长时期根和叶中的欧前胡素含量 Fig. 2 Content of imperatorin in roots and leaves at different growth stages |
2.2 AdPAL1基因的组织特异性表达分析
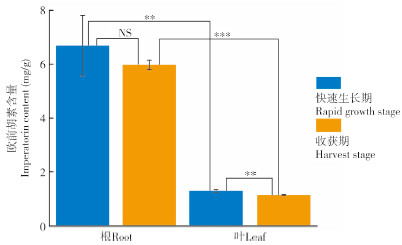
取禹白芷快速生长期和收获期的根和叶,通过qRT-PCR分析AdPAL1基因在不同组织中的表达情况。结果(图 3)表明,快速生长期根中AdPAL1基因的表达量显著高于叶,约是叶中表达量的21倍;收获期根中AdPAL1基因的表达量约为叶中的50.21%。此外,根中AdPAL1基因表达变化量与欧前胡素累积变化量的皮尔逊相关系数为0.46(中度相关),而叶中AdPAL1基因表达变化量与欧前胡素累积变化量的皮尔逊相关系数为0.17(弱相关),综合以上研究结果,初步判断在根中AdPAL1基因参与欧前胡素的生物合成。
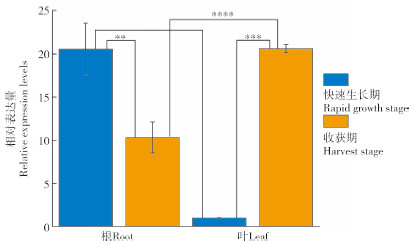
|
| **、*** 分别表示0.01、0.001水平上差异显著;NS表示差异不显著 **, *** indicate significant differences at 0.01, 0.001 levels, respectively; NS indicates that the difference is not significant 图 3 不同生长时期根和叶中的AdPAL1基因相对表达 Fig. 3 Relative expression of AdPAL1 gene in roots and leaves at different growth stages |
2.3 禹白芷AdPAL1基因序列克隆
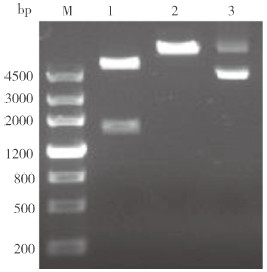
以禹白芷幼叶cDNA为模板,使用特异性引物扩增出约2 000 bp的片段(图 4)。通过TA克隆获得重组质粒pMD19-AdPAL1,酶切检测结果显示双酶切获得的2个DNA片段大小之和等于单酶切片段大小,表明AdPAL1基因已成功插入到pMD19T载体中。
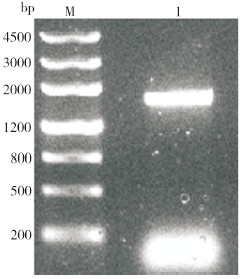
|
| M:Marker,DNA分子量标准;1:AdPAL1基因扩增结果 M: Marker, DNA standard; 1: AdPAL1 gene amplif ication result 图 4 AdPAL1基因的PCR扩增结果 Fig. 4 PCR amplification result of AdPAL1 gene |
2.4 AdPAL1蛋白生物信息学分析
利用Genetyx和DNAMAN软件对AdPAL1基因ORF序列进行分析,结果表明AdPAL1基因ORF长1 764 bp,编码557个氨基酸。本研究中所克隆到的AdPAL1基因与二代转录组拼接获得的AdPAL1核苷酸序列并不完全一致,二者相似性为99.64%。将该序列上传至GenBank数据库,获得GenBank登录号为OQ822236。
使用GenBank数据库中的BLASTp程序对AdPAL1蛋白进行比对分析,结果发现禹白芷AdPAL1与石防风属植物KpPAL2蛋白序列相似性最高(99.28%),与紫苏(Perilla frutescens)PfPAL蛋白相似性稍低(88.31%)。利用DNAMAN将AdPAL1蛋白序列与从NCBI中挑选的相似性较高的部分序列进行多序列比对分析,结果表明AdPAL1蛋白与已知蛋白相似性很高,在第39~54位氨基酸残基之间含有保守基序“GTITASGDLVPLSYIA”和Ala-Ser-Gly催化三联体,这被预测为典型的苯丙氨酸和组氨酸解氨酶(HAL)特征模式[22-23](图 5)。此外,AdPAL1蛋白还包含PAL保守催化活性位点,如GLALVNG(96~102)、NDN(223~225)和HNQDV(327~331)[8, 24](图 5)。利用IQTREE2将AdPAL1蛋白序列与已发表文献中相似性较高的序列进行系统发育分析,结果进化树分为4个进化枝,分别为单子叶植物、苔藓植物、裸子植物和双子叶植物,其中禹白芷AdPAL1蛋白与伞形科植物当归、欧芹、野胡萝卜、紫花前胡、石防风属植物的PAL蛋白聚为同一大进化枝,尤其是与石防风属植物KpPAL2蛋白处于同一小进化枝(图 6),表明二者亲缘关系较近,具有相似或相同的结构与功能。
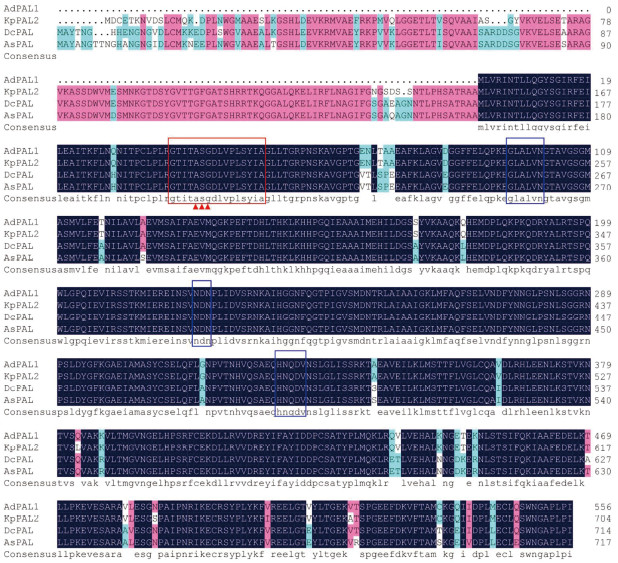
|
| 黑色:相似性=100%;粉红色:75% ≤相似性<100%;浅蓝色:50% ≤相似性<75%;保守的基序使用红框标记,保守的活性位点基序使用三角形标记,催化活性位点使用蓝框标记 Black: Similarity = 100%; Pink: 75% ≤ Similarity < 100%; Light blue: 50% ≤ Similarity < 75%; Conservative motif, active site motif, catalytic active site are marked with a red box, a triangle, and a blue box, respectively 图 5 AdPAL1蛋白与其他植物PAL蛋白的多序列比对结果 Fig. 5 Multiple sequence alignment results of AdPAL1 protein with other plant PAL proteins |
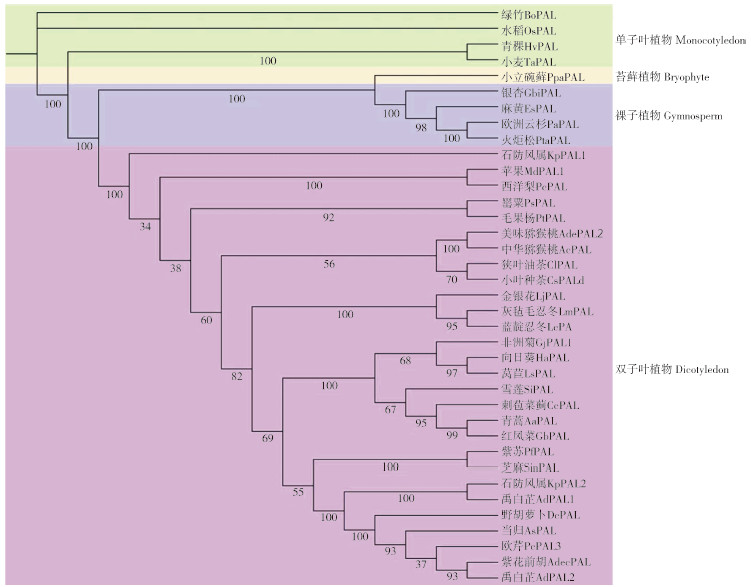
|
| 图 6 AdPAL1蛋白与其他植物中PAL蛋白的系统发育分析 Fig. 6 Phylogenetic analysis of AdPAL1 protein and other plant PAL proteins |
使用ExPASy ProtParam tool对AdPAL1蛋白进行理化性质分析,结果表明AdPAL1蛋白单体相对分子质量为61.10 kD,理论等电点(pI)为5.92;带正电氨基酸残基(Arg + Lys)为62,带负电氨基酸残基(Asp + Glu)为54。蛋白不稳定指数为36.77,属于稳定蛋白;脂肪指数为93.68,总平均疏水性(GRAVY)为-0.124,为亲水蛋白。AdPAL1蛋白含有20种基本氨基酸,其中亮氨酸含量最高、为11.10%;其次是丙氨酸和谷氨酸,分别为7.90% 和7.50%;色氨酸含量最低、为0.4%。
利用SSpro方法对AdPAL1蛋白进行二级结构分析,结果发现该蛋白二级结构中α- 螺旋(H)占58.53%,无规则卷曲(C)占38.78%,延伸带(E)占2.69%。利用Swiss-Model Workspace使用自动模式预测AdPAL1蛋白的三级结构,发现该模型为同源四聚体(图 7),是以欧芹苯丙氨酸解氨酶[6f6t.1.A]为模板,在第1~557位氨基酸处建模,序列相似度为94.23%。使用SMART软件进行蛋白保守结构域预测,结果表明AdPAL1蛋白在第1~338氨基酸位置具有芳香族氨基酸裂解酶结构域。使用InterPro在线工具对AdPAL1蛋白进行蛋白家族归类分析,结果表明AdPAL1蛋白属于芳香族氨基酸裂解酶(IPR001106,1~556)家族、苯丙氨酸解氨酶(IPR005922,1~547)亚家族成员。
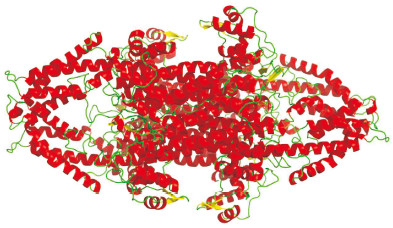
|
| 红色:α-螺旋;黄色:β-折叠;绿色:环 Red: α-spiral; Yellow: β-fold; Green: Ring 图 7 AdPAL1蛋白四聚体的三维结构预测 Fig. 7 3D Structure prediction of AdPAL1 protein tetramer |
使用SignalP 6.0服务器分析AdPAL1蛋白,未发现信号肽,表明该蛋白为非分泌型蛋白。利用TMHMM工具预测AdPAL1蛋白的跨膜螺旋区,结果表明AdPAL1蛋白不含跨膜螺旋区域,为非膜蛋白。利用亚细胞定位软件ProtComp 9.0预测AdPAL1蛋白,结果显示其定位于细胞质。
蛋白相互作用预测结果表明,AdPAL1蛋白与C4H、4CL1、4CL2、4CL3、4CL5、HISB6B、AAS、ACOS5(类4CL酶)、CCoAMT1、AT4G05160等10个蛋白存在互作,其中PAL、C4H、4CL、CCoAMT是简单香豆素生物合成的催化酶,AAS参与细胞氨基酸代谢,AT4G05160参与脂肪酸的激活,HISN6B为参与组氨酸生物合成的组氨酸磷酸转氨酶(图 8)。
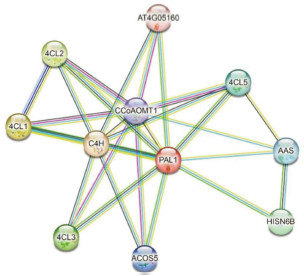
|
| C4H:反式肉桂酸4-单加氧酶;4CL1、4CL2、4CL3、4CL5:4-香豆酸CoA连接酶;ACOS5:类4-香豆酸CoA连接酶;HISB6B:组氨酸磷酸氨基转移酶;AAS:芳香醛合酶;CCoAMT1:咖啡酰CoA 3-O- 甲基转移酶;AT4G05160:AMP依赖性合成酶和连接酶家族蛋白 C4H: Trans cinnamic acid 4-monooxygenase; 4CL1, 4CL2, 4CL3, 4CL5:4-coumaric acid CoA ligase; ACOS5: 4-coumaric acid CoA ligase like; HISB6B: Histidine phosphate aminotransferase; AAS: Aromatic aldehyde synthase; CCoAMT1: Coffee acyl CoA 3-O-methyltransferase; AT4G05160:AMP dependent synthase and ligase family proteins 图 8 AdPAL1蛋白互作网络预测结果 Fig. 8 AdPAL1 protein interaction network prediction results |
2.5 AdPAL1基因原核表达载体构建
使用NdeI和HindIII双酶切质粒ProS2- AdPAL1,可切出目的片段和载体,且两片段之和等于单酶切片段长度(图 9),表明AdPAL1基因已成功插入原核表达载体ProS2中。
|
| M:Marker,DNA分子量标准;1~2:质粒ProS2-AdPAL1的NdeI、HindIII双酶切和HindIII单酶切;3:质粒对照 M: Marker, DNA standard; 1: NdeI and HindIII double digestion; 2: HindIII single digestion results of plasmid ProS2-AdPAL1; 3: plasmid control 图 9 质粒ProS2-AdPAL1酶切检测结果 Fig. 9 Enzyme digestion detection results of plasmid ProS2-AdPAL1 |
2.6 AdPAL1基因原核表达
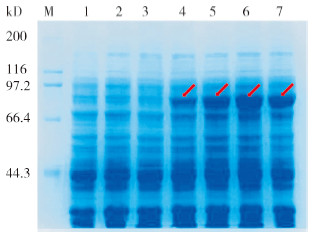
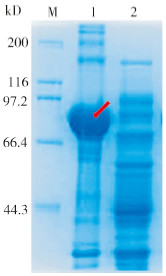
将重组质粒ProS2-AdPAL1转化大肠杆菌BL21(DE3)后,使用IPTG进行诱导表达。在15 ℃、终浓度为0.5 mmol/L IPTG下,分别诱导表达0、2、4、6、8 h后,提取细菌总蛋白进行SDS-PAGE分析。结果表明,与对照相比,ProS2-AdPAL1转化菌经IPTG诱导后,在相对分子质量84 kD(含Protein S标签蛋白23 kD)左右有1条蛋白条带,并且随诱导时间增加其蛋白含量逐渐增加,表明重组质粒ProS2-AdPAL1在大肠杆菌BL21(DE3)中成功诱导表达了AdPAL1蛋白。当温度为15 ℃、诱导时间为6 h时,蛋白表达量最高(图 10)。为检测AdPAL1融合蛋白的存在形式,对诱导表达8 h的菌体进行超声破碎,然后使用SDS-PAGE检测,结果表明AdPAL1蛋白全部以包涵体形式存在(图 11)。
|
| M:Marker,蛋白分子量标准;1~2:15 ℃、IPTG终浓度为0.5 mmol/L下ProS2转化菌分别诱导0和8 h的总蛋白;3~7:相同条件下ProS2-AdPAL1转化菌分别诱导0、2、4、6和8 h的总蛋白 M: Marker, protein standard; 1-2: Total proteins of ProS2 transformed bacteria induced for 0 and 8 h under 0.5 mmol/L of IPTG at 15 ℃; 3-7: Total proteins of ProS2-AdPAL1 transformed bacteria induced for 0, 2, 4, 6, and 8 h under the same conditions 图 10 15 ℃下不同诱导时间对AdPAL1蛋白表达的影响 Fig. 10 Effects of different induction times on AdPAL1 protein expression at 15 ℃ |
|
| M:Marker,蛋白分子量标准;1:细胞破碎后的沉淀;2:细胞破碎后的上清 M: Marker, protein standard; 1: Precipitate after cell disruption; 2: Supernatant after cell disruption 图 11 AdPAL1重组蛋白存在形式的检测 Fig. 11 Detection of the presence form of AdPAL1 recombinant protein |
3 讨论
植物PAL在介导和协调初级到次级代谢的碳通量方面发挥着关键作用,这对于植物应对不同的环境条件以适应和优化生长至关重要[25]。禹白芷的主要药效成分为香豆素类化合物,通过苯丙素途径合成[3]。在植物中,PAL是苯丙素途径的第一个限速酶[26]。因此,禹白芷AdPAL基因的表达情况直接或间接影响香豆素的生物合成。本研究中,不同生长时期禹白芷根和叶中欧前胡素变化量与AdPAL1基因表达变化量具有中度相关性,判断AdPAL1基因可能参与欧前胡素的生物合成。因此,对禹白芷AdPAL1基因进行克隆和生物信息学分析,结果表明所克隆的AdPAL1基因ORF全长1 764 bp,编码557个氨基酸,pI为5.92,其与番红花CsPAL蛋白(559 aa,pI为5.7)类似,但比白花前胡PpPAL1(718 aa,pI为6.25)和PpPAL2蛋白(705 aa,pI为6.16)5′端要短[8],比孜然CcPAL蛋白(116 aa,pI为5.85)要长得多[13]。蛋白保守结构域预测结果显示,AdPAL1蛋白在第1~338位氨基酸位置具有芳香族氨基酸裂解酶结构域。InterPro软件则将AdPAL1蛋白归类为芳香族氨基酸裂解酶(IPR001106,1~556)家族、苯丙氨酸解氨酶(IPR005922,1~547)亚家族成员。事实上,芳香族氨基酸裂解酶家族包含苯丙氨酸解氨酶(PAL,EC4.3.1.24)、组氨酸解氨酶(HAL,EC24.3.1.3)和酪氨酸氨基变位酶(EC5.4.3.6)三类[27-29]。PAL和HAL是类裂合酶I超家族成员,均催化类似的β消除反应,形成同源四聚体才具有活性,且二者均具有翻译后环化特征及催化作用的Ala-Ser-Gly三位一体结构[30]。PAL是植物和真菌中苯丙素组装的关键生物合成催化剂,并参与多种次生代谢产物如黄烷类、呋喃香豆素类植保素及细胞壁成分的生物合成。这些化合物对于植物正常生长和环境胁迫响应至关重要。HAL催化组氨酸降解的第一步,即从组氨酸中去除氨基团以产生尿刊酸。PAL和HAL中的核心结构域约有30% 的序列一致性,其中PAL包含从共同折叠处延伸的约160个残基[31]。酪氨酸2, 3- 氨基突变酶具有氨基突变酶活性,并且在较小程度上具有氨裂解酶活性[32]。这表明AdPAL1为苯丙氨酸解氨酶。近期研究发现,石防风属植物KpPAL2能够催化从L- 苯丙氨酸到反式肉桂酸的反应[8]。本研究中,AdPAL1蛋白与KpPAL2蛋白序列相似性高达78.44%,并与KpPAL2处于同一进化枝,这进一步表明AdPAL1为苯丙氨酸解氨酶。因此,本研究所克隆基因为PAL基因。
在植物中,蛋白的亚细胞定位对于基因功能的解析至关重要。在本研究中,预测结果显示AdPAL1蛋白定位于细胞质,这与白花前胡中PpPAL蛋白定位于细胞质的结果相一致[7]。PAL基因的表达也具有组织和时空特异性,并与活性化合物的生物合成相关联。在前胡中,MeJA、UV和冷处理诱导的PpPAL基因表达与白花前胡甲素含量呈正相关[7]。在巴戟天中,McPAL3在叶片和成熟果实中表达,McPAL2仅在叶片中表达,McPAL1主要在果实幼龄期表达,但仅有McPAL3的表达模式与东莨菪内酯含量累积规律一致[33]。半夏的药用部位为块茎,PtPAL基因在叶中表达量最高,其次是块茎和根,花中表达量最低[34]。冬凌草的药用部位为茎和叶,IrPAL基因在叶片中表达量最高,茎次之,根中表达量最低[35]。本研究中,禹白芷的药用部位为根,且快速生长期的AdPAL1基因主要在根中表达,收获期在根中的表达量下降约50%,这暗示AdPAL1基因参与禹白芷欧前胡素的生物合成。此外,收获期禹白芷叶片中AdPAL1基因的表达量显著高于快速生长期,这与巴戟天中老叶McPAL3基因的表达量远远高于幼叶和成熟叶的现象一致[33]。这可能是老叶中乙烯含量升高[36],并诱导AdPAL1和McPAL3基因上调表达的结果[33]。下一步将对AdPAL1蛋白进行纯化和酶活分析,为进一步阐明禹白芷欧前胡素生物合成途径奠定基础。
4 结论本研究发现,根中AdPAL1基因表达的变化与欧前胡素含量的变化呈中度相关,初步证明AdPAL1基因参与了欧前胡素的生物合成。成功克隆AdPAL1基因,该基因ORF长1 764 bp,编码557个氨基酸,单体相对分子质量为61.10 kD,理论等电点为5.92。AdPAL1蛋白为亲水稳定蛋白,定位于细胞质,无信号肽、跨膜螺旋区域,其二级结构主要由α- 螺旋和无规则卷曲构成,三级结构呈同源四聚体形式。AdPAL1与石防风属植物KpPAL2蛋白亲缘关系最近,且可能与香豆素生物合成途径中的C4H、4CL、CCoAMT等蛋白存在相互作用。构建原核表达载体ProS2- AdPAL1,并在大肠杆菌BL21(DE3)中成功表达出AdPAL1蛋白,表达形式为包涵体。本研究结果对于揭示AdPAL1基因的功能和欧前胡素生物合成途径具有重要参考价值。
| [1] |
国家药典委员会. 中国药典[M]. 北京: 中国医药科技出版社, 2020: 109. National Pharmacopoeia Commission. Chinese pharmacopoeia[M]. Beijing: China Medical Science and Technology Press, 2020: 109. |
| [2] |
LIU Q, LU Z, HE W, LI F, CHEN W, LI C, CHAO Z, TIAN E. Development and characterization of 16 novel microsatellite markers by transcriptome sequencing for Angelica dahurica and test for crossspecies amplification[J]. BMC Plant Biology, 2020, 20(1): 152. DOI:10.1186/s12870-020-02374-8 |
| [3] |
ZHAO L, ZHANG S, SHAN C, SHI Y, WU H, WU J, PENG D. De novo transcriptome assembly of Angelica dahurica and characterization of coumarin biosynthesis pathway genes[J]. Gene, 2021, 791: 145713. DOI:10.1016/j.gene.2021.145713 |
| [4] |
JUN S Y, SATTLER S A, CORTEZ G S, VERMERRIS W, SATTLER S E, KANG C. Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase[J]. Plant Physiology, 2017, 176(2): 1452-1468. DOI:10.1104/pp.17.01608 |
| [5] |
KHAKDAN F, ALIZADEH H, RANJBAR M. Molecular cloning, functional characterization and expression of a drought inducible phenylalanine ammonia-lyase gene (ObPAL) from Ocimum basilicum L[J]. Plant Physiology and Biochemistry, 2018, 130: 464-472. DOI:10.1016/j.plaphy.2018.07.026 |
| [6] |
RAES J, ROHDE A, CHRISTENSEN J H, VAN DE PEER Y, BOERJAN W. Genome-wide characterization of the lignification toolbox in Arabidopsis[J]. Plant Physiology, 2003, 133(3): 1051-1071. DOI:10.1104/pp.103.026484 |
| [7] |
SUI Z, LUO J, YAO R, HUANG C, ZHAO Y, KONG L. Functional characterization and correlation analysis of phenylalanine ammonialyase (PAL) in coumarin biosynthesis from Peucedanum praeruptorum Dunn[J]. Phytochemistry, 2019, 158: 35-45. DOI:10.1016/j.phytochem.2018.11.006 |
| [8] |
TONG Z, XIE J, YIN M, WU J, ZHA L, CHU S, PENG H. Cloning, characterization and prokaryotic expression analysis of two phenylalanine ammonia-lyase genes from Peucedanum praeruptorum Dunn[J]. Brazilian Journal of Botany, 2022, 45: 897-907. DOI:10.1007/s40415-022-00826-z |
| [9] |
HUANG X, BAI X, XIE Z, FAHAD S, GBOKIE T, LU Y, GUO T, LI J, ZHANG Z, WU W, YI K. De novo transcriptome assembly of Coffea liberica reveals phylogeny and expression atlas of phenylalanine ammonia-lyase genes in Coffea species[J]. Industrial Crops and Products, 2023, 192: 116029. DOI:10.1016/j.indcrop.2022.116029 |
| [10] |
ZHU B F, LIU Y, PEI X Q, WU Z L. Characterization of phenylalanine ammonia lyases from lettuce (Lactuca sativa L.) as robust biocatalysts for the production of D-and L -amino acids[J]. Journal of Agricultural and Food Chemistry, 2023, 71(6): 2935-2942. DOI:10.1021/acs.jafc.2c07890 |
| [11] |
MO F, LI L, ZHANG C, YANG C, CHEN G, NIU Y, SI J, LIU T, SUN X, WANG S, WANG D, CHEN Q, CHEN Y. Genome-wide analysis and expression profiling of the phenylalanine ammonia-lyase gene family in Solanum tuberosum[J]. International Journal of Molecular Sciences, 2022, 23(12): 6833. DOI:10.3390/ijms23126833 |
| [12] |
TORK S D, NAGY E Z A, TOMOIAGĂ R B, BENCZE L C. Engineered, scalable production of optically pure l-phenylalanines using phenylalanine ammonia-lyase from Arabidopsis thaliana[J]. The Journal of Organic Chemistry, 2023, 88(2): 852-862. DOI:10.1021/acs.joc.2c02106 |
| [13] |
HABIBOLLAHI M, KAVOUSI H R, LOHRASBI-NEJAD A, RAHPEYMA S A. Cloning, characterization and expression of a phenylalanine ammonia-lyase gene (CcPAL) from cumin (Cuminum cyminum L.)[J]. Journal of Applied Research on Medicinal and Aromatic Plants, 2020, 18: 100253. DOI:10.1016/j.jarmap.2020.100253 |
| [14] |
IM J H, YU H W, PARK C H, KIM J W, SHIN J H, JANG K Y, PARK Y J. Phenylalanine ammonia-lyase: A key gene for color discrimination of edible mushroom Flammulina velutipes[J]. Journal of Fungi, 2023, 9(3): 339. DOI:10.3390/jof9030339 |
| [15] |
GHALAMBORAN M R, KOHNAVARD A, HASSANI S B. Phenylalanine response in rice kernel under chitosan nanoparticles spraying[J]. Acta Physiologiae Plantarum, 2023, 45(4): 61. DOI:10.1007/s11738-023-03538-3 |
| [16] |
WANG H, WANG X, ZHOU L, ZHANG S, AN L, BAO J, LI Z, SUN Y, LI Y, CUI J, JIN D Q, ZHANG J, XU J, GUO Y. Structural characteristics and in vitro and in vivo immunoregulatory properties of a gluco-arabinan from Angelica dahurica[J]. International Journal of Biological Macromolecules, 2021, 183: 90-100. DOI:10.1016/j.ijbiomac.2021.04.077 |
| [17] |
陈琳, 唐志书, 宋忠兴, 刘妍如, 胡锦航, 史鑫波, 孙琛, 江大海, 李晓红. 不同产地白芷药材9个呋喃香豆素成分的含量测定及其质量评价[J]. 中国中药杂志, 2019, 44(14): 3002-3009. DOI:10.19540/j.cnki.cjcmm.20190505.102 CHEN L, TANG Z S, SONG Z X, LIU Y R, HU J H, SHI X B, SUN C, JIANG D H, LI X H. Quantitative determination of nine furanocoumarins for quality evaluation of Angelica dahurica from different habitats[J]. China Journal of Chinese Materia Medica, 2019, 44(14): 3002-3009. DOI:10.19540/j.cnki.cjcmm.20190505.102 |
| [18] |
GUO J, HU Z, YAN F, LEI S, LI T, LI X, XU C, SUN B, PAN C, CHEN L. Angelica dahurica promoted angiogenesis and accelerated wound healing in db/db mice via the HIF-1α/PDGF-β signaling pathway[J]. Free Radical Biology and Medicine, 2020, 160: 447-457. DOI:10.1016/j.freeradbiomed.2020.08.015 |
| [19] |
张玲, 李彩霞, 张海晨, 马娜, 杨娇, 张晓东. 滇龙胆异戊烯基焦磷酸异构酶基因的克隆与表达分析[J]. 广东农业科学, 2015, 42(17): 139-146. DOI:10.16768/j.issn.1004-874X.2015.17.003 ZHANG L, LI C X, ZHANG H C, MA N, YANG J, ZHANG X D. Cloning and expression analysis of isopentenyl pyrophosphate isomerase gene in Gentiana rigescens[J]. Guangdong Agricutural Sciences, 2015, 42(17): 139-146. DOI:10.16768/j.issn.1004-874X.2015.17.003 |
| [20] |
贾俊婷, 陈兵先, 吴柔贤, 戴彰言, 钟明生, 解昊, 刘双幸, 刘军. 水稻组蛋白去甲基化酶基因OsJMJ719的克隆及表达分析[J]. 广东农业科学, 2022, 49(11): 152-161. DOI:10.16768/j.issn.1004-874X.2022.11.016 JIA J T, CHEN B X, WU R X, DAI Z Y, ZHONG M S, XIE H, LIU S X, LIU J. Analysis on cloning and expression of histone demethylase gene OsJMJ719 in rice[J]. Guangdong Agricultural Sciences, 2022, 49(11): 152-161. DOI:10.16768/j.issn.1004-874X.2022.11.016 |
| [21] |
HE Y, ZHONG Y, BAO Z, WANG W, XU X, GAI Y, WU J. Evaluation of Angelica decursiva reference genes under various stimuli for RTqPCR data normalization[J]. Scientific Reports, 2021, 11(1): 18993. DOI:10.1038/s41598-021-98434-6 |
| [22] |
HE Y, ZHONG X, JIANG X, CONG H, SUN H, QIAO F. Characterisation, expression and functional analysis of PAL gene family in Cephalotaxus hainanensis[J]. Plant Physiology and Biochemistry, 2020, 156: 461-470. DOI:10.1016/j.plaphy.2020.09.030 |
| [23] |
MA W, WU M, WU Y, REN Z, ZHONG Y. Cloning and characterisation of a phenylalanine ammonia-lyase gene from Rhus chinensis[J]. Plant Cell Reports, 2013, 32(8): 1179-1190. DOI:10.1007/s00299-013-1413-6 |
| [24] |
MA R F, LIU Q Z, XIAO Y, ZHANG L, LI Q, YIN J, CHEN W S. The phenylalanine ammonia-lyase gene family in Isatis indigotica Fort.: molecular cloning, characterization, and expression analysis[J]. Chinese Journal of Natural Medicines, 2016, 14(11): 801-812. DOI:10.1016/S1875-5364(16)30097-8 |
| [25] |
BARROS J, DIXON R A. Plant phenylalanine/tyrosine ammonia-lyases[J]. Trends in Plant Science, 2020, 25(1): 66-79. DOI:10.1016/j.tplants.2019.09.011 |
| [26] |
BARBER M S, MITCHELL H J. Regulation of phenylpropanoid metabolism in relation to lignin biosynthesis in plants[J]. International Review of Cytology, 1997, 172: 243-293. DOI:10.1016/S0074-7696(08)62362-1 |
| [27] |
SCHWEDE T F, RéTEY J, SCHUL Z G E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile[J]. Biochemistry, 1999, 38(17): 5355-5361. DOI:10.1021/bi982929q |
| [28] |
RACHID S, KRUG D, KUNZE B, KOCHEMS I, SCHARFE M, ZABRISKIE T M, BLöCKER H, MüLLER R. Molecular and biochemical studies of chondramide formation—Highly cytotoxic natural products from Chondromyces crocatus Cmc5[J]. Chemistry & Biology, 2006, 13(6): 667-681. DOI:10.1016/j.chembiol.2006.06.002 |
| [29] |
APPERT C, LOGEMANN E, HAHLBROCK K, SCHMID J, AMRHEIN N. Structural and catalytic proper ties of the four phenylalanine ammonia‐lyase isoenzymes from parsley (Petroselinum crispum Nym.)[J]. European Journal of Biochemistry, 1994, 225(1): 491-499. DOI:10.1111/j.1432-1033.1994.00491.x |
| [30] |
PILBáK S, TOMIN A, RéTEY J, POPPE L. The essential tyrosinecontaining loop conformation and the role of the C‐terminal multihelix region in eukaryotic phenylalanine ammonia‐lyases[J]. FEBS Journal, 2006, 273(5): 1004-1019. DOI:10.1111/j.1742-4658.2006.05127.x |
| [31] |
CALABRESE J C, JORDAN D B, BOODHOO A, SARIASLANI S, VANNELLI T. Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis[J]. Biochemistry, 2004, 43(36): 11403-11416. DOI:10.1021/bi049053+ |
| [32] |
KRUG D, MüLLER R. Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF[J]. ChemBioChem, 2009, 10(4): 741-750. DOI:10.1002/cbic.200800748 |
| [33] |
WANG Q, LIU J, YANG F, JIA D, WU T. The phenyla lanine ammonia-lyase gene McPAL3: the key gene involved in the scopoletin accumulation of Morinda citrifolia L.[J]. Brazilian Journal of Botany, 2021, 44(3): 663-670. DOI:10.1007/s40415-021-00717-9 |
| [34] |
何潇, 刘兴, 辛正琦, 谢海艳, 辛余凤, 吴能表. 半夏PtPAL基因的克隆, 表达与酶动力学分析[J]. 作物学报, 2021, 47(10): 1941-1952. DOI:10.3724/SP.J.1006.2021.04227 HE X, LIU X, XIN Z Q, XIE H Y, XIN Y F, WU N B. Molecular cloning, expression, and enzyme kinetic analysis of a phenylalanine ammonia-lyase gene from Pinellia ternate[J]. Acta Agronomica Sinica, 2021, 47(10): 1941-1952. DOI:10.3724/SP.J.1006.2021.04227 |
| [35] |
黄进勇, 张济萌, 郭瀚师, 王璐, 岳彩鹏, 张靖楠. 冬凌草PAL基因的克隆及表达分析[J]. 郑州大学学报(理学版), 2020, 52(3): 115-119. DOI:10.13705/j.issn.1671-6841.2019443 HUANG J Y, ZHANG J M, GUO H S, WANG L, YUE C P, ZHANG J N. Cloning and expression analysis of PAL gene from Isodon rubescens[J]. Journal of Zhengzhou University (Natural Science Edition), 2020, 52(3): 115-119. DOI:10.13705/j.issn.1671-6841.2019443 |
| [36] |
MERCHANTE C, ALONSO J M, STEPANOVA A N. Ethylene signaling: simple ligand, complex regulation[J]. Current Opinion in Plant Biology, 2013, 16(5): 554-560. DOI:10.1016/j.pbi.2013.08.001 |
(责任编辑 马春敏)
 2023, Vol. 50
2023, Vol. 50