文章信息
基金项目
- 江西省教育厅科学技术研究项目(GJJ211739,GJJ201720);上饶市科技局项目(2021年)
作者简介
-
乐翔兆,博士,讲师,上饶师范学院生命科学学院专任教师,主要从事昆虫生理生化与分子生物学研究,曾首次在基因组水平揭示黑腹果蝇几丁质合成酶A基因(kkv)的转录调控机制。近5年先后主持和参与江西省教育厅科学技术研究项目等3项,江西省昆虫学会会员,在《 Insect Science》《 PLOS Biology》《 Frontiers in Physiology》等国内外期刊发表学术论文6篇,参与编写专著《婺源醉美自然保护地》。
乐翔兆(1991—),女,博士,讲师,研究方向为昆虫几丁质合成通路基因功能及转录调控机制,E-mail:yuexiangzhaosrsy@163.com.
文章历史
- 收稿日期:2023-12-05
害虫严重危害农业生产以及人类健康,而虫害的防治很大程度上依赖于杀虫剂的使用。由于昆虫种群中杀虫剂耐药性的迅速发展,害虫防控面临的挑战逐渐增加,寻找新的害虫防治靶点用于开发新型杀虫剂成为研究热点[1-2]。几丁质是N-乙酰氨基葡萄糖(N-acetylglucosamine, GlcNAc)残基通过β-1, 4糖苷键连接而成的线性聚合物,是自然界中储存量第二的天然多糖,仅次于纤维素。目前发现几丁质广泛存在于真菌、昆虫和甲壳类动物等生物体中,是昆虫外骨骼、气管及中肠围食膜的重要组分[3]。昆虫体内富含几丁质的结构在保护其抵御物理伤害、化学毒性和环境中的病原微生物侵染等方面起着至关重要的作用[3-5]。由于几丁质结构在昆虫中是必不可少的,且不存在于植物和脊椎动物中,因此成为防控害虫的理想靶标。
为满足生长发育需要,昆虫定期蜕皮过程中,外骨骼、气管及中肠围食膜中富含几丁质的角质层需要替换为新的、更为宽松的角质层来保障虫体扩张所需的空间。因此,几丁质的合成在昆虫每个发育阶段均受到动态调控[6]。昆虫体内几丁质生物合成通路,始于海藻糖,终于几丁质,共有8个酶参与,包括海藻糖酶(Trehalase,TRE)、己糖激酶(Hexokinase,HK)、谷氨酰胺: 果糖-6-磷酸转氨酶(Glutamine: fructose-6-phosphate aminotransferase,GFAT)、UDP-N-乙酰葡糖胺焦磷酸化酶(UDP-N-acetylglucosamine pyrophosphorylase,UAP)和几丁质合成酶(Chitin synthase,CHS)[7]。CHS是几丁质生物合成通路上最后一种酶,催化几丁质合成。在部分昆虫中仅发现存在1种CHS,而多数昆虫中则存在2种[8]。昆虫CHS分为CHS1(或称CHSA)和CHS2(或称CHSB)两类,CHS1基因在外骨骼和气管中大量表达,催化几丁质合成,而CHS2基因则主要在中肠围食膜表达。这两类CHS在昆虫的变态和摄食行为中发挥着不同作用[8-12]。在昆虫胚胎期破坏CHS1功能可导致虫体严重的发育障碍,表皮角质层发育不完整,头骨退化,背气管干发育异常,局部膨大或者收缩,导致胚胎死亡[9,12-13]。对昆虫若虫或者幼虫CHS1进行RNA干扰(RNA interference),往往导致虫体出现严重的蜕皮障碍,气管内螺旋变薄,虫体大量死亡[10,12,14-15]。在一些昆虫中,CHS1存在可变剪切体,某些CHS1可变剪切的mRNA下调可引起其他缺陷表型。例如,家蚕(Bombyx mori)蛹中期翅盘中表达的CHS1-2b被敲低引起翅发育功能障碍[16]。此外,昆虫中肠表达的CHS2基因受到抑制后,会导致中肠变短,围食膜结构被破坏,昆虫体重下降,甚至因饥饿而死亡[14,17-18]。
几丁质合成酶在合成几丁质过程中发挥至关重要的作用,为认识调控CHS基因表达变化的因子,现就CHS基因的转录调控研究进展进行综述,可为进一步研究破坏昆虫几丁质合成通路、开发昆虫防治技术新靶点提供理论基础。
1 昆虫激素及其相关基因对几丁质合成酶的转录调控有报道显示,多种昆虫的CHS转录受调控蜕皮与变态的内源激素蜕皮激素(20-Hydroxyecdysone,20E)及保幼激素(Juvenile hormone,JH)的影响(表 1)。20E/ 蜕皮激素受体(Ecdysone receptor,EcR)/ 超气门蛋白(Ultraspiracle,USP)复合物上调一系列与蜕皮相关的早期应答基因,例如Broad Complex(BR-C)、Ecdysone-induced protein 75(E75)以及E74[20-22]。这些早期表达的转录因子随后激活晚期应答基因的表达,如Ftz transcription factor 1(Ftz-F1)、Hormone receptor 3(HR3)和E78B[23]。

|
在黑腹果蝇(Drosophila metamorphosis)3龄幼虫和化蛹后不久的虫体中,几乎无法检测到CHS mRNA。然而,在第1次20E脉冲反应中,CHS1 mRNA首先上调,与蛹的内表皮形成同时发生;CHS2 mRNA则在几小时后上调,与蛹的前表皮形成同时发生。果蝇CHS1启动子区域中预测存在BR-C结合位点,而在CHS2启动子区域预测存在BR-C和E74A结合位点,但这2个基因启动子区域预测结果显示并无EcR和USP结合位点[24]。Tellam等[25]在分析果蝇CHS1基因潜在的启动子序列时,则发现其中存在类似20E应答元件的序列。Gangishetti等[26]研究发现,果蝇表皮过表达转录因子Grainy head(Grh)可引起CHS1转录水平上调,且体外实验证明一段含有预测的Grh结合位点的CHS1基因启动子DNA可与Grh蛋白结合。Grh抑制编码20E生物合成途径最后一种酶的基因Shade(Shd)的表达,且调控一些20E应答基因包括Ftz-F1的表达,而果蝇Shd突变体Shd7C94胚胎晚期CHS1的转录水平有所下调[26-27]。
激素调控昆虫CHS转录的研究主要集中在鳞翅目昆虫中。烟草天蛾(Manduca sexta)CHS1被认为受到20E负调控[28]。烟草天蛾5龄幼虫摄食期间CHS1表达水平相对稳定,但在摄食停止后其表达水平急剧下降,并在徘徊期再次逐渐上升,在蛹蜕皮时达到高峰,相反地,20E在徘徊期前升高,在化蛹前降低[29-31]。斜纹夜蛾(Spodoptera exigua)CHS1和CHS2表达受外源注射的20E诱导,以及双链RNA(Double-stranded RNA, dsRNA)dsEcR的抑制[32]。与果蝇CHS启动子区域中的预测结果相似,在亚洲玉米螟(Ostrinia furnacalis)CHS2的核心启动子区中,也同样发现存在潜在的BR-C和E74A结合位点,并未预测发现存在EcR和USP结合位点[33]。亚洲玉米螟5龄幼虫被注射20E后的24 h内,CHS1各个可变剪切体mRNA表达均上调[15]。20E和蜕皮激素激动剂RH5992被报道可引起6龄云山卷叶蛾(Choristoneura fumiferana)蜕皮异常及死亡,且均抑制CHS1表达;一定浓度的外源JH类似物甲氧普烯(Methoprene)则可诱导CHS1的表达[34]。粘虫(Mythimna separata)被注射20E后6 h和12 h,CHS2基因表达上调[35]。家蚕蜕皮后内源20E水平较低而JH水平较高时,外源20E和甲氧普烯均可显著上调CHS2的表达量;在蜕皮期,当内源20E水平较高而JH水平较低时,外源20E对CHS2表达的促进作用并不明显,甲氧普烯对CHS2表达的促进作用几乎可忽略不计;在家蚕取食桑叶时,内源激素水平较低,CHS2在甲氧普烯的作用下显著上调[36]。张捷等[37]研究发现,在注射外源20E的家蚕中,CHS1-2a的表达被抑制,而在注射甲氧普烯后CHS1-2a只在48 h轻微上调。此外,在家蚕中敲低正调控JH生物合成的POUM2基因抑制了CHS1和CHS2表达[38]。Zhang等[39]发现在家蚕CHS1-2b启动子区域存在1个Fox顺式调控元件,可与FoxJ蛋白结合,在蛹期增强CHS1-2b的转录;而在蛹前期,20E诱导的BR-C Z4抑制FoxJ转录,间接抑制CHS1-2b转录。然而,舞毒蛾(Lymantria dispar)BR-C被敲低后,CHS1和CHS2表达均下调,表明BR-C对CHS表达具有促进作用[40]。美国白蛾(Hyphantria cunea)幼虫的E74被敲低显著降低了CHS1和CHS2基因的表达[41]。
赤拟谷盗(Tribolium castaneum)HR3可正调控多个几丁质代谢基因的转录,其敲低也引起了CHS1轻微下调,但下调并不显著 [42]。东亚飞蝗(Locusta migratoria)HR3被敲低后,CHS1基因则显著下调[43]。此外,东亚飞蝗中,涉及蜕皮激素生物合成的CYP307a2基因被敲低引起了CHS1上调[44]。有研究发现在马铃薯甲虫(Leptinotarsa decemlineata)CHS1可变剪切CHS1a、CHS1b和CHS2的mRNA水平与循环周期性变化的20E和JH峰值一致。中肠体外培养和活体内生物测定结果均显示CHS的3种mRNA表达可被外源20E诱导。对马铃薯甲虫幼虫Shd基因进行RNA干扰,降低了这3种mRNA水平。此外,通过对EcR、E75、HR3以及Ftz-F1进行RNA干扰从而对20E信号进行干扰,均减少了这些CHS基因的转录。蜕皮激素激动剂氯虫酰肼(Halofenozide)则刺激了这3种CHS表达;体外培养和活体内生物实验结果则表明外源JH和甲氧普烯也可激活这3种CHS表达,而敲低幼虫JH生物合成基因JH酸甲基转移酶(JH acid methyltransferase,JHAMT)则降低了这些CHS基因的mRNA水平[45]。
上述研究表明昆虫体内CHS转录受到激素20E与JH的共同调控,但20E和JH对CHS1转录的调控作用在不同报道中并不完全一致。20E可能通过20E/EcR/USP调控CHS基因的转录,但这种调控作用并不是通过其与CHS基因启动子直接结合而实现的,而是可能由下游应答基因BR-C、FoxJ、E75、Ftz-F1以及HR3介导间接产生的作用。
2 昆虫表皮及中肠所受刺激对几丁质合成酶转录的影响转录因子Grh在昆虫表皮屏障形成及上皮修复过程中发挥作用。在受伤的果蝇胚胎中,酪氨酸激酶Cadherin 96Ca(Cad96ca)被激活,并正调控Grh。德国小蠊(Blattella germanica)中,Grh和Cad96ca的下调均引起了蜕皮障碍,且导致CHS1基因表达上调,CHS1的上调被认为可能是由HR3表达上调介导的[46]。在果蝇表皮受损后数分钟之内,CHS1的转录在表皮伤口处被激活(表 1)。果蝇CHS1第1个内含子被预测具有转录因子Grh与AP-1-like共识位点,且含该位点的CHS1内含子DNA片段具有表皮伤口处增强子的功能,但并非由Grh或AP-1-like激发该增强子在伤口处的活性[47]。昆虫中肠围食膜中,几丁质对维持肠道功能具有重要作用,多项研究表明围食膜中CHS2的表达受到昆虫进食的影响。Ibrahim等[48]发现埃及伊蚊(Aedes aegypti)中肠CHS2表达量在吸血后增加。在亚洲玉米螟4龄和5龄幼虫取食期,虫体内CHS2 mRNA表达量也有所增加[33]。东方果实蝇(Bactrocera dorsalis)的CHS2在发育各阶段中肠中均有表达,但在摄食期表达量较高[49]。家蚕CHS2的表达也在蜕皮结束后随着摄食逐渐增加[36]。东亚飞蝗CHS2在若虫和成虫的摄食期均有表达,且飞蝗饥饿处理后CHS2的表达被显著抑制,重新进食后,CHS2表达量则快速上调[18,50]。然而,饲喂5龄欧洲玉米螟(Ostrinia nubilalis)幼虫后,对其进行24 h饥饿处理,CHS2的转录水平显著增加[17]。以上研究结果均表明昆虫CHS2表达水平受进食情况影响(表 1)。
可见,与器官的功能相对应,昆虫表皮受损伤后可能通过诱导CHS表达促进伤口愈合,但在此情况下同样被诱导表达的Grh并不是CHS上调的原因,中肠围食膜CHS的表达则受到进食情况影响。
3 昆虫糖代谢相关因子对几丁质合成酶的转录调控昆虫几丁质合成通路的起始物质为海藻糖,其主要在脂肪体中通过海藻糖-6-磷酸合成酶(Trehalose-6-phosphate synthase,TPS)及海藻糖磷酸脂酶(Trehalose-6-phosphate phosphatase,TPP)合成。TPS催化葡萄糖-6-磷酸与UDP- 葡萄糖生成海藻糖-6-磷酸,然后通过TPP最终合成海藻糖。海藻糖在有能量需求时则通过TRE降解为葡萄糖,用于能量供给[7,51]。
昆虫几丁质合成通路的第1个酶为TRE,其在多项研究中被证明可间接调控CHS的mRNA表达(表 1)。在甜菜夜蛾(Spodoptera exigua)幼虫发育过程中,敲低3龄幼虫编码可溶性海藻糖酶的TRE1基因或编码膜结合海藻糖酶的TRE2基因,CHS1和CHS2的转录均受到抑制,但TRE1下调对CHS1影响较大,而TRE2下调对CHS2影响较大[52]。Tang等[53]发现赤拟谷盗5个海藻化酶基因,特别是TRE1-4和TRE2,都可通过调控几丁质生物合成途径中的基因表达而导致蜕皮畸形和高死亡率。在赤拟谷盗被分别注射5种海藻酶的dsRNA后48 h,CHS1a和CHS1b mRNA相对表达量显著下降,CHS2表达水平在注射dsTRE1-2、dsTRE1-3或dsTRE2后48 h也显著下降,但在注射dsTRE1-4后48 h显著上升。Zhao等[54]发现褐飞虱(Nilaparvata lugens)被注射dsTRE1-1或dsTREs(含dsTRE1-1、dsTRE1-2和dsTRE2)后72 h,CHS1表达水平显著降低,但张露等[55]报道褐飞虱在被注射dsTRE1(含dsTRE1-1、dsTRE1-2)后表皮CHS1a表达量上升,注射dsTRE1-2后CHS1的表达量上升。褐飞虱中TRE被敲低后还可引起翅芽CHS1b表达量显著降低,TRE抑制剂有效霉素(Validamycin)也可以抑制翅芽和表皮中CHS1基因的表达[55-56]。异色瓢虫(Harmonia axyridi)TRE2被敲低后,CHS1及CHS2的表达均显著下降[57]。受到有效霉素的影响,柑橘木虱(Diaphorina citri)CHS1及粘虫CHS2基因的表达被显著抑制,东方果实蝇CHS1及CHS2的表达均被抑制[35,58-59]。除TRE外,其他部分几丁质合成通路基因也影响CHS基因表达。褐飞虱HK被敲低引起CHS1、CHS1a和CHS1b表达量降低[60]。褐飞虱、美国白蛾的GFAT基因下调后,引起CHS1下调[61-62]。甜菜夜蛾UAP被敲低后,CHS2表达明显下降[63]。上述研究均表明几丁质合成通路基因TRE、HK、GFAT和UAP基因表达量的下调普遍抑制了CHS基因的转录,仅在赤拟谷盗及褐飞虱相关的少数研究中发现TRE基因被敲低后引起CHS转录水平上调的情况[53,55]。
几丁质合成通路基因之外的一些几丁质代谢基因也可调控CHS表达(表 1)。敲低白背飞虱(Sogatella furcifera)编码几丁质去乙酰酶(Chitin deacetylases,CDA)的基因CDA1、CDA2、CDA4或几丁质酶基因(Chitinase,Cht)Cht 5、Cht7、Cht10及IDGF2后,均可引起CHS1、CHS1a及CHS1b显著下调[64-65]。敲低烟草甲(Lasioderma serricorne)CDA1也抑制了CHS1表达[66]。草地贪夜蛾(Spodoptera frugiperda)Cht被敲低则抑制了CHS2表达[67]。暗黑鳃金龟(Holotrichia parallela)中参与几丁质降解的β-N-乙酰氨基葡萄糖酶基因(β-N-acetylglucosaminidase,NAG)和N-乙酰氨基葡萄糖激酶基因(N-acetylglucosamine kinase,NAGK)被敲低均引起CHS下调[68]。
在多种昆虫中,海藻糖合成通路的TPS可间接影响CHS转录(表 1)。柑橘大实蝇(Bactrocera minax)幼虫被注射dsTPS后,其TPS酶活性和海藻糖含量显著下降,包括CHS1基因在内的几丁质合成通路3个关键基因mRNA表达下调[69]。在褐飞虱中注射dsTPS1或者dsTPS2,其海藻糖含量均有所增加,CHS1a的mRNA水平在注射dsTPS1或dsTPS2后48 h时降低、72 h时回升,CHS1b的mRNA水平仅在注射dsTPS1后48 h时降低,72 h时回升[70]。Zhou等[71]也研究发现抑制褐飞虱TPS的表达影响海藻糖和几丁质的代谢,引起几丁质含量的下降,对褐飞虱TPS1、TPS2以及新发现的TPS3进行RNA干扰,注射dsTPS3后72 h, CHS1表达量显著下调,而在注射dsTPSs后48、72 h,CHS1、CHS1a以及CHS1b表达水平先下降后上升。赤拟谷盗注射dsTPS-1和dsTPS-2后48、72 h,CHS2 mRNA下调,而CHS1 mRNA仅在注射dsTPS-1后48 h有轻微下调,在注射dsTPS-2后72 h则显著上调,部分昆虫无法正常化蛹,几丁质含量下降[72]。豌豆蚜(Acyrthosiphon pisum Harris)TPS基因被敲低后,几丁质含量下降,且在注射dsTPS后24~48 h,多个几丁质代谢相关基因(包括CHS2)表达被抑制[73]。烟粉虱(Bemisia tabaci)中TPS2被敲低后也出现了较为显著的CHS表达量下调情况[74]。粘虫体内TPS被敲低后,中肠及表皮的几丁质含量均显著降低,CHS1和CHS2 mRNA相对表达量显著改变,CHS1在12 h下调,但在24、72 h上调,而CHS2在72 h上调,96 h下调[75]。白背飞虱在被注射dsTPS1和dsTPS2后几丁质含量显著上升,CHS1(CHS1a及CHS1b)与CHS1a相对表达量在注射dsTPS1或dsTPS2后均极显著上升,且注射dsTPS1和dsTPS2混合的dsTPSs后CHS1b的表达也极显著增加[76]。
葡萄糖代谢是昆虫糖代谢的重要过程,糖原对维持昆虫体内海藻糖水平稳定非常重要,一些相关基因也可影响CHS1转录(表 1)。磷酸糖脲酶(Phosphoglucomutase,PGM)是昆虫糖酵解和糖异生途径的关键酶,调节糖原和海藻糖的代谢,敲低PGM1或PGM2基因均可抑制褐飞虱CHS1(含CHS1a和CHS1b)的表达[77]。糖原合酶激酶(Glycogen synthase kinase, GSK-3)是位于糖代谢途径中的关键酶,其可通过胰岛素信号调节体内能量代谢过程。褐飞虱被注射dsGSK-3后48 h,CHS1a表达极显著增加而CHS1b表达显著降低;注射dsGSK-3和葡萄糖的混合物后48 h,CHS1表达未发生显著变化,而dsGSK-3和海藻糖的混合物对CHS1的表达有明显抑制作用,但各处理组中CHS1的表达在72 h均被抑制[78]。磷酸果糖激酶(Phosphofructokinase,PFK)是糖酵解途径中的主要限速酶,其活性显著影响葡萄糖的消耗和能量产生[79]。褐飞虱中,注射dsPFK引起CHS1a和CHS1b上调[61]。果糖1, 6-二磷酸(Fructose 1, 6-diphosphate,FDP)是糖酵解途径中的内源性中间体,也是PFK的变构激活剂。外源变构剂FDP-Na处理美国白蛾幼虫引起CHS1基因表达显著下调,而CHS2显著上调[80]。
上述研究表明糖代谢相关基因及物质,包括诸多涉及海藻糖合成通路、几丁质合成与降解、葡萄糖代谢的基因,对CHS表达存在复杂的调控作用,其具体调控机制有待进一步研究。
4 MicroRNA对昆虫几丁质合成酶mRNA表达水平的影响MicroRNA(miRNA)为一类由内源基因编码的、长度约为22 nt的非编码单链小RNA分子,在动植物中参与转录后基因表达调控[81-82]。在飞蝗CHS1的编码区存在miR-71的功能性结合位点,若虫蜕皮过程中,miR-71负调控CHS1 mRNA[81]。褐飞虱CHS1a的3'UTR区域预测存在miR-2703的结合位点,在HEK293T和S2细胞中,miR-2703可负调控褐飞虱CHS1a,且20E应答基因BR-C编码的转录因子可抑制miR-2703的表达,而miR-2703和CHS1a的转录水平在褐飞虱2~5龄若虫发育过程中表达模式相反[82]。白纹伊蚊(Aedes albopictus)中,miR-989可促进CHS1表达,并且编码1种几丁质结合蛋白的基因XM_029863591.1(被证实为miR-989的靶基因),敲低XM_029863591.1也引起了CHS1显著上调[83]。这些研究表明CHS1 mRNA受到miRNA的调控(表 1),miR-71、miR-2703可作用于CHS1 mRNA实现对CHS1的负调控,而miR-989对CHS1转录水平的影响则是通过间接作用实现的。
5 几丁质合成抑制剂对昆虫几丁质合成酶的转录调控苯甲酰基脲类杀虫剂是一类特异性昆虫生长调节剂,具有触杀和胃毒作用,可干扰幼虫及卵内胚胎发育过程中的几丁质合成[84]。第一种通过抑制昆虫几丁质形成而起作用的苯甲酰脲类商业杀虫剂名为二氟脲(Diflubenzuron),其被发现能抑制丝氨酸蛋白酶,这可能会阻断几丁质合成酶转化为活性的酶,从而阻碍几丁质合成,并因此干扰昆虫蜕皮以及引起其他生理变化[85-86]。二氟脲处理四斑按蚊(Anopheles quadrimaculatus)幼虫后,CHS1表达水平显著升高,而虫体的几丁质含量随着处理组二氟脲浓度升高而逐渐下降[85]。Lu等[86]研究发现,柑橘木虱在暴露于二氟脲后24~48 h,CHS1表达显著提高,且出现蜕皮异常以及成虫翅卷曲的表型。Zhang等[87]研究发现,随着二氟脲浓度的增加,柑橘木虱CHS1表达量呈上升趋势,且蜕皮失败率和虫体死亡率均呈上升趋势。Yao等[88]用二氟脲处理爻纹细蛾(Conopomorpha sinensis)的卵,引起CHS1、CHS2表达量上调。这些研究均表明二氟脲可诱导昆虫CHS1转录水平上调(表 1)。
除二氟脲外,苯甲酰基脲类农药氟虫脲(Flufenoxuron)、氟啶脲(Chlorfluazuron)等也可影响昆虫CHS转录(表 1)。刘晓健等[89]研究发现,氟虫脲浸渍处理后的中华稻蝗(Oxya chinensis)及东亚飞蝗若虫CHS1表达量提高。Chang等[90]用含有虱螨脲(Lufenuron)的饲料饲喂辣椒实蝇(Bactrocera latifrons),雄性和雌性成虫的CHS2表达均上调。陈金鹏等[91]用化合物HN-21和虱螨脲分别对草地贪夜蛾幼虫进行处理,发现2种处理均引起CHS1及CHS2基因表达下调。在经过亚致死浓度(60 mg/L)氟啶脲处理后,棉铃虫(Helicoverpa armigera)CHS1基因表达呈现出“抑制-激活-再抑制”的波动趋势[92]。
其他非苯甲酰基脲类的几丁质合成抑制也同样被报道可影响CHS转录(表 1)。几丁质合成抑制剂尼可霉素Z(Nikkomycin Z)是几丁质合成酶底物UDP-乙酰葡糖胺(UDP-GlcNAc)的类似物,能够不可逆地抑制几丁质合成酶的活性[93]。当家蚕暴露于尼可霉素Z中,蜕皮幼虫的中肠围食膜中几丁质含量减少,而在蜕皮末期和刚蜕皮的幼虫中检测到CHS2表达量异常升高[36]。暴露于杂环类几丁质合成抑制剂噻嗪酮(Buprofezin)的棉蚜(Aphis gossypii)F0代成虫CHS1 mRNA相对表达量显著增加,暴露于亚致死浓度LC 15(1.125 mg/L)和LC30(2.888 mg/L)后,F1代幼虫和成虫期CHS1 mRNA的相对表达量也显著增加[94]。
这些研究证明上述几丁质合成抑制剂可调控CHS表达,但产生的影响并不完全一致。二氟脲、氟虫脲、尼可霉素Z及噻嗪酮的使用均导致昆虫CHS mRNA相对表达量增加,HN-21及虱螨脲则抑制了草地贪夜蛾CHS的表达,而虱螨脲可促进辣椒实蝇CHS2表达,氟啶脲对棉铃虫CHS1的影响则呈波动性变化。
6 其他影响几丁质合成酶转录的因素已报道的调控CHS转录的转录因子多与20E应答相关,但也有研究鉴定出其他参与调控CHS转录的转录因子,且尚无证据显示这些转录因子对CHS转录的影响与20E相关(表 1)。Zhao等[95]发现,甜菜夜蛾Fox被敲低后,引起幼虫白化且部分蜕皮畸形,CHS1 mRNA表达水平在注射dsFox 24 h后显著下降,而在72 h后转而升高,CHS2 mRNA水平则在36 h后显著下降。果蝇S2细胞中全基因组RNAi筛选出537个对CHS1启动子活性有影响的基因,其中Akirin、NFAT、48 related 3(Fer3)、Autophagyrelated 101(Atg101)被全身敲低,可引起3龄幼虫CHS1基因启动子活性下调,且体外实验证明bHLH家族转录因子Fer3可通过与果蝇CHS1启动子特异性结合促进CHS1的转录[96]。DNA甲基化也可调节CHS1转录,DNA甲基化抑制剂5-氮杂胞苷-2'-脱氧胞苷(5-azacytidine-2'-deoxycytidine,5-aza-dC)处理家蚕后,蛹表皮基因启动子的甲基化率降低,CHS1-2b转录显著增加;蛹特异性转录因子Deformed epidermal autoregulatory factor-1(DEAF1)与未甲基化的基因内的启动子结合,激活了蛹中翅的CHS1-2b转录。DNA甲基转移酶1(DNA Methyltransferase 1,DNMT1)和DEAF1通过竞争性结合启动子中的CpG岛来影响CHS1-2b的转录[97]。此外,赤拟谷盗中有机物萘和苯被发现可增加CHS2 mRNA的表达水平[98-99]。一种源于白刀豆(Canavalia ensiformis)的脲酶异构体Jaburetox(长度约为11 kD的重组肽),被昆虫食用后有致命毒性。长红锥蝽(Rhodnius prolixus)被饲喂Jaburetox溶液18 h后,中枢神经系统和唾液腺的CHS mRNA表达水平下降[100]。上述研究表明CHS转录受到多方面因子的调控,显示了CHS转录调控机制的复杂性。
7 展望在害虫防控当中,由于害虫抗药性的产生,商业化杀虫剂的功效随着时间推移逐渐减弱[101]。为了避免因害虫抗药性导致的害虫暴发,研发作用于新靶点的新型化合物非常重要。昆虫几丁质转录水平表达受到多种内因和外因的调控,包括昆虫激素、表皮损伤及进食等应激刺激、糖代谢相关因子、几丁质合成抑制剂、miRNA等,且这些因子间也存在复杂的互作关系(表 1、图 1)。因此,尽管对几丁质合成酶基因表达或蛋白活性进行干扰可以引起昆虫大量死亡,但虫体内为应对几丁质合成异常、补偿CHS功能的减弱,形成相应的反馈调节机制。几丁质合成通路基因TRE、HK、UAP及GFAT表达的下调,普遍可抑制CHS基因转录[54,56-63]。因此,CHS表达量或酶活性的下降,引起上游底物的积累,可能促进CHS转录。在抑制CHS基因表达或酶活性的同时,可通过抑制几丁质合成上游底物的积累,例如使用海藻糖酶抑制剂,有效降低昆虫几丁质的合成从而防控害虫。已报道的海藻糖酶抑制剂除有效霉素外,还包括海藻唑啉(Trehazolin)、Salbostatin、2R, 5R-二羟甲基、3R, 4R-二羟基吡咯等,但这些抑制剂在昆虫活体中抑制海藻糖酶的效果欠佳[51,102]。因此,仍有待进一步研究发掘靶作用于几丁质合成通路CHS上游的其他基因或物质的有效抑制剂。
|
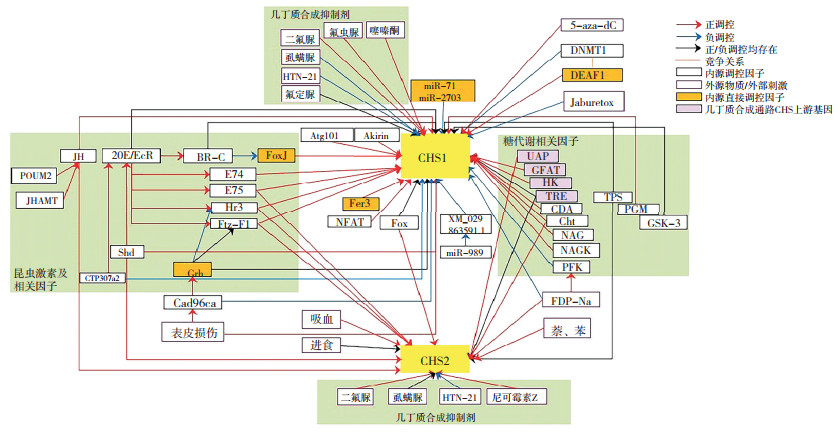
| 图 1 昆虫几丁质合成酶转录调控网络 Fig. 1 Transcriptional regulatory network of insect chitin synthases |
此外,转录的RNA也可能引发机体反馈调节,以维持内部稳态。近年来,有研究报道转录的RNA会对基因转录产生反馈调控。基因表达受转录因子控制,转录因子由DNA结合域和激活域组成。Boija等[103]报道了多种转录因子可以通过其激活域相分离能力与转录中介体复合物Mediator相互作用,发生液-液相分离,激活基因表达。Henninger等[104]提出转录形成的RNA可通过调控Mediator的相分离从而对转录过程进行反馈调节的机制。尽管多种转录因子可调控CHS转录,但目前关于直接调控CHS转录的报道较少。转录因子中,仅有Grh、FoxJ、DEAF1和Fer3被报道可与CHS1启动子结合调控CHS1转录,直接调控因子miR-71及miR-2703则通过作用于CHS1的非启动子区,在转录后调控mRNA的水平(图 1)。CHS基因是否也存在转录的RNA引发的反馈调节机制仍待进一步研究。
虽然大量研究表明CHS表达受激素、糖类物质等多种因子的间接调控,但详细的调控机制研究仅主要围绕20E及其应答因子对CHS1的转录调控,其他因子对CHS转录的详细调控模式和机制尚不明确,尤其是对CHS2基因的转录调控机制研究更少(图 1)。已有的研究结果为进一步深入研究CHS转录的具体机制提供了一定基础,但各种间接调控因子间可能存在复杂的相互作用,这可能为进一步研究CHS转录调控机制带来一定干扰,但高通量测序技术的发展,以及基因、蛋白等互作信息的不断丰富,使得探究CHS转录的具体调控机制及分析其调控网络成为可能。进一步深入研究CHS转录的分子调控机制,探究其直接调控因子,以及探究上游间接调控因子是如何具体地影响CHS转录,特别是CHS转录的具体反馈调节机制,从而发掘新的害虫防控靶标,可为进一步加强以CHS为靶标的害虫控制手段提供新思路。
| [1] |
REN Y, LI Y, JU Y, ZHANG W, WANG Y. Insect cuticle and insecticide development[J]. Archives of Insect Biochemistry and Physiology, 2023, 114(4): e22057. DOI:10.1002/arch.22057 |
| [2] |
陈金翠, 曹利军, 马中正, 苑新新, 宫亚军, 沈修婧, 魏书军. 不同龄期草地贪夜蛾对常用杀虫剂的敏感性[J]. 广东农业科学, 2022, 49(8): 81-86. DOI:10.16768/j.issn.1004-874X.2022.08.010 CHEN J C, CAO L J, MA Z Z, YUAN X X, GONG Y J, SHEN X J, WEI S J. Susceptibility of different instar larvae of Spodoptera frugiperda to commonly used insecticides[J]. Guangdong Agricultural Sciences, 2022, 49(8): 81-86. DOI:10.16768/j.issn.1004-874X.2022.08.010 |
| [3] |
安沙, 刘文娟, 张忠, 张瑞玲. 基于RNAi的昆虫几丁质合成酶类调控研究进展[J]. 中华卫生杀虫药械, 2022, 28(4): 372-378. DOI:10.19821/j.1671-2781.2022.04.022 AN S, LIU W J, ZHANG Z, ZHANG R L. Researches on the regulation of insect chitin synthesis enzymes based on RNA interference[J]. Chinese Journal of Hygienic Insecticides and Equipment, 2022, 28(4): 372-378. DOI:10.19821/j.1671-2781.2022.04.022 |
| [4] |
张道伟, 钱正敏, 张正玲, 骆颖. 二斑叶螨几丁质酶基因的克隆及特性分析[J]. 广东农业科学, 2015, 42(12): 128-134, 4. DOI:10.16768/j.issn.1004-874X.2015.12.014 ZHANG D W, QIAN Z M, ZHANG Z L, LUO Y. Cloning and characteristics analysis of a chitinase gene from Tetranychus urticae Koch[J]. Guangdong Agricultural Sciences, 2015, 42(12): 128-134, 4. DOI:10.16768/j.issn.1004-874X.2015.12.014 |
| [5] |
MUTHUKRISHNAN S, MERZENDORFER H, ARAKANE Y, KRAMER K J. 7-chitin metabolism in insects//Gilbert LI. Lnsect molecular biology and biochemistry[M]. San Diego: Academic Press, 2012: 193-235.
|
| [6] |
ZHU K Y, MERZENDORFER H, ZHANG W, ZHANG J, MUTHUKRISHNAN S. Biosynthesis, turnover, and functions of chitin in insects[J]. Annual Review of Entomology, 2016, 61: 177-196. DOI:10.1146/annurev-ento-010715-023933 |
| [7] |
张文庆, 陈晓菲, 唐斌, 田宏刚, 陈洁, 姚琼. 昆虫几丁质合成及其调控研究前沿[J]. 应用昆虫学报, 2011, 48(3): 475-479. ZHANG W Q, CHEN X F, TANG B, TIAN H G, CHEN J, YAO Q. Insect chitin biosynthesis and its regulation[J]. Chinese Journal of Applied Entomology, 2011, 48(03): 475-479. |
| [8] |
许静静, 李思琪, 任梦圆, 薛雨欣, 李永强. 昆虫几丁质合成关键酶功能及其RNAi技术在害虫防治中的研究进展[J]. 陕西农业科学, 2022, 68(8): 1-10. XU J J, LI S Q, REN M Y, XUE Y X, LI Y Q. Advance of research in function of key enzymes involving in chitin synthesis and their application combined with RNAi technique in pest control[J]. Shaanxi Journal of Agricultural Sciences, 2022, 68(8): 1-10. |
| [9] |
MOUSSIAN B, LETIZIA A, MARTÍNEZ-CORRALES G, ROTSTEIN B, CASALI A, LLIMARGAS M. Deciphering the genetic programme triggering timely and spatially-regulated chitin deposition[J]. PLoS Genetics, 2015, 11(3): e1005054. DOI:10.1371/journal.pgen.1004939 |
| [10] |
JIANG L H, MU L L, JIN L, ANJUM A A, LI G Q. RNAi for chitin synthase 1 rather than 2 causes growth delay and molting defect in Henosepilachna vigintioctopunctata[J]. Pesticide Biochemistry & Physiology, 2021, 178: 104934. DOI:10.1016/j.pestbp.2021.104934 |
| [11] |
WANG Y, FAN H W, HUANG H J, XUE J, WU W J, BAO Y Y, XU H J, ZHU Z R, CHENG J A, ZHANG C X. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae)[J]. Insect Biochemistry & Molecular Biology, 2012, 42(9): 637-646. DOI:10.1016/j.ibmb.2012.04.009 |
| [12] |
OSTROWSKI S, DIERICK H A, BEJSOVEC A. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster[J]. Genetics, 2002, 161(1): 171-182. DOI:10.1093/genetics/161.1.171 |
| [13] |
MOUSSIAN B, SCHWARZ H, BARTOSZEWSKI S, NÜSSLEINVOLHARD C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster[J]. Journal of Morphology, 2005, 264(1): 117-130. DOI:10.1002/jmor.10324 |
| [14] |
ARAKANE Y, MUTHUKRISHNAN S, KRAMER KJ, SPECHT C A, TOMOYASU Y, LORENZEN M D, KANOST M, BEEMAN R W. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix[J]. Insect Biology, 2005, 14(5): 453-463. DOI:10.1111/j.1365-2583.2005.00576.x |
| [15] |
QU M, YANG Q. Physiological significance of alternatively spliced exon combinations of the single-copy gene class A chitin synthase in the insect Ostrinia furnacalis (Lepidoptera)[J]. Insect Molecular Biology, 2012, 21(4): 395-404. DOI:10.1111/j.1365-2583.2012.01145.x |
| [16] |
XU G, ZHANG J, LYU H, LIU J, DING Y, FENG Q, SONG Q, ZHENG S. BmCHSA-2b, a Lepidoptera specific alternative splicing variant of epidermal chitin synthase, is required for pupal wing development in Bombyx mori[J]. Insect Biochemistry & Molecular Biology, 2017, 87: 117-126. DOI:10.1016/j.ibmb.2017.06.017 |
| [17] |
KHAJURIA C, BUSCHMAN L L, CHEN M S, MUTHUKRISHNAN S, ZHU K Y. A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae[J]. Insect Biochemistry & Molecular Biology, 2010, 40(8): 621-629. DOI:10.1016/j.ibmb.2010.06.003 |
| [18] |
LIU X, ZHANG H, LI S, ZHU K Y, MA E, ZHANG J. Characterization of a midgut-specific chitin synthase gene (LmCHS2) responsible for biosynthesis of chitin of peritrophic matrix in Locusta migratoria[J]. Insect Biochemistry & Molecular Biology, 2012, 42(12): 902-910. DOI:10.1016/j.ibmb.2012.09.002 |
| [19] |
ZHANG C, DING Y, ZHOU M, TANG Y, CHEN R, CHEN Y, WEN Y, WANG S. RNAi-mediated CHS-2 silencing affects the synthesis of chitin and the formation of the peritrophic membrane in the midgut of Aedes albopictus larvae[J]. Parasit Vectors, 2023, 16(1): 259. DOI:10.1186/s13071-023-05865-3 |
| [20] |
DIBELLO P R, WITHERS D A, BAYER C A, FRISTROM J W, Guild G M. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers[J]. Genetics, 1991, 129(2): 385-397. DOI:10.1093/genetics/129.2.385 |
| [21] |
SEGRAVES W A, HOGNESS D S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily[J]. Genes & Development, 1990, 4(2): 204-219. DOI:10.1101/gad.4.2.204 |
| [22] |
BURTIS K C, THUMMEL C S, JONES C W, KARIM F D, HOGNESS D S. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins[J]. Cell, 1990, 61(1): 85-99. DOI:10.1016/0092-8674(90)90217-3 |
| [23] |
THUMMEL C S. Molecular mechanisms of developmental timing in C. elegans and Drosophila[J]. Developmental Cell, 2001, 1: 453-465. DOI:10.1016/s1534-5807(01)00060-0 |
| [24] |
GAGOU M E, KAPSETAKI M, TURBERG A, KAFETZOPOULOS D. Stage-specific expression of the chitin synthase DmeChSA and DmeChSB genes during the onset of Drosophila metamorphosis[J]. Insect Biochemistry & Molecular Biology, 2002, 32(2): 141-146. DOI:10.1016/s0965-1748(01)00101-1 |
| [25] |
TELLAM R L, VUOCOLO T, JOHNSON S E, JARMEY J, PEARSON R D. Insect chitin synthase cDNA sequence, gene organization and expression[J]. European Journal of Biochemistry, 2000, 267(19): 6025-6043. DOI:10.1046/j.1432-1327.2000.01679.x |
| [26] |
GANGISHETTI U, VEERKAMP J, BEZDAN D, SCHWARZ H, LOHMANN I, MOUSSIAN B. The transcription factor Grainy head and the steroid hormone ecdysone cooperate during differentiation of the skin of Drosophila melanogaster[J]. Insect Molecular Biology, 2012, 21(3): 283-295. DOI:10.1111/j.1365-2583.2012.01134.x |
| [27] |
PETRYK A, WARREN J T, MARQUÉS G, JARCHO M P, GILBERT L I, KAHLER J, PARVY J P, LI Y, DAUPHIN-VILLEMANT C, O'CONNOR M B. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone[J]. Proceedings of the National Academy of Sciences, 2003, 100(24): 13773-13778. DOI:10.1073/pnas.2336088100 |
| [28] |
MERZENDORFER H, ZIMOCH L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases[J]. Journal of Experimental Biology, 2003, 206(24): 4393-4412. DOI:10.1242/jeb.00709 |
| [29] |
ZHU Y C, SPECHT C A, DITTMER N T, MUTHUKRISHNAN S, KANOST M R, KRAMER K J. Sequence of a cDNA and expression of the gene encoding a putative epidermal chitin synthase of Manduca sexta[J]. Insect Biochemistry & Molecular Biology, 2002, 32(11): 1497-1506. DOI:10.1016/s0965-1748(02)00070-x |
| [30] |
BAKER F C, TSAI L W, REUTER C C, SCHOOLEY D A. In vivo fluctuation of JH, JH acid, and ecdysteroid titer, and JH esterase activity during development of fifth stadium Manduca sexta[J]. Insect Biochemistry, 1987, 17(7): 989-996. DOI:10.1016/0020-1790(87)90108-9 |
| [31] |
BOLLENBACHER W E, SMITH S L, GOODMAN W, GILBERT L I. Ecdysteroid titer during larval-pupal-adult development of the tobacco hornworm, Manduca sexta[J]. General and Comparative Endocrinolog y, 1981, 44(3): 302-306. DOI:10.1016/0016-6480(81)90005-8 |
| [32] |
YAO Q, ZHANG D, TANG B, CHEN J, CHEN J, LU L, ZHANG W. Identification of 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway[J]. PLoS One, 2010, 5(11): e14058. DOI:10.1371/journal.pone.0014058 |
| [33] |
QU M, LIU T, YANG J, YANG Q. The gene, expression pattern and subcellular localization of chitin synthase B from the insect Ostrinia furnacalis[J]. Biochemical & Biophysical Research Communications, 2011, 404(1): 302-307. DOI:10.1016/j.bbrc.2010.11.111 |
| [34] |
AMPASALA D R, ZHENG S, ZHANG D, LADD T, DOUCET D, KRELL P J, RETNAKARAN A, FENG Q. An epidermis-specific chitin synthase cDNA in Choristoneura fumiferana: Cloning, characterization, developmental and hormonal-regulated expression[J]. Archives of Insect Biochemistry & Physiology, 2011, 76(2): 83-96. DOI:10.1002/arch.20404 |
| [35] |
王晓曦, 赵桂莹, 杨航, 张雅男, 樊东. 蜕皮激素和有效霉素对粘虫几丁质合成酶B基因表达的影响[J]. 应用昆虫学报, 2019, 56(5): 1026-1036. DOI:10.7679/j.issn.2095-1353.2019.113 WANG X X, ZHAO G Y, YANG H, ZHANG Y N, FAN D. Effect of ecdysone and validamycin on chitin synthase B gene expression in Mythimna separate[J]. Chinese Journal of Applied Entomology, 2019, 56(5): 1026-1036. DOI:10.7679/j.issn.2095-1353.2019.113 |
| [36] |
ZHUO W, CHU F, KONG L, TAO H, SIMA Y, XU S. Chitin synthase B: A midgut-specific gene induced by insect hormones and involved in food intake in Bombyx mori larvae[J]. Archives of Insect Biochemistry & Physiology, 2014, 85(1): 36-47. DOI:10.1002/arch.21141 |
| [37] |
张捷, 丁洋, 郑思春. 家蚕表皮型几丁质合成酶基因BmCHSA-2a的鉴定及其对激素的响应[J]. 华南师范大学学报: 自然科学版, 2018, 50(1): 65-71. DOI:10.6054/j.jscnun.2018039 ZHANG J, DING Y, ZHENG S. Identification of epidermal chitin synthase BmCHSA-2a in Bombyx mori and its responses to hormones[J]. Journal of South China Normal University (Natural Science Edition), 2018, 50(1): 65-71. DOI:10.6054/j.jscnun.2018039 |
| [38] |
CAI R, CHEN X, YANG W, WANG X, SUN L, ZHAO P, XIA Q, HE H, WANG Y. POUM2 homeostasis regulates intimal remodeling and cells fate in the anterior silk gland of the silkworm[J]. International Journal of Biological Macromolecules, 2023, 225: 715-729. DOI:10.1016/j.ijbiomac.2022.11.135 |
| [39] |
ZHANG J, XU G, QIU B, ZHANG X, FENG Q, YANG Q, ZHENG S. BR-C Z4 and FoxJ interact to regulate expression of a chitin synthase gene CHSA-2b in the pupal wing discs of the silkworm, Bombyx mori[J]. Insect Biochemistry & Molecular Biology, 2020, 116: 103264. DOI:10.1016/j.ibmb.2019.103264 |
| [40] |
DING N, WANG Z, GENG N, ZOU H, ZHANG G, CAO C, LI X, ZOU C. Silencing Br-C impairs larval development and chitin synthesis in Lymantria dispar larvae[J]. Journal of Insect Physiology, 2020, 122: 104041. DOI:10.1016/j.jinsphys.2020.104041 |
| [41] |
ZHANG B, YAO B, LI X, JING T, ZHANG S, ZOU H, ZHANG G, ZOU C. E74 knockdown represses larval development and chitin synthesis in Hyphantria cunea[J]. Pesticide Biochemistry and Physiology, 2022, 187: 105216. DOI:10.1016/j.pestbp.2022.105216 |
| [42] |
KIM B E, CHOI B, PARK W R, KIM Y J, MUN S, CHOI H S, KIM D K. Nuclear receptor HR3 mediates transcriptional regulation of chitin metabolic genes during molting in Tribolium castaneum[J]. Pest Management Science, 2022, 78(10): 4377-4387. DOI:10.1002/ps.7056 |
| [43] |
ZHAO X, QIN Z, LIU W, LIU X, MOUSSIAN B, MA E, LI S, ZHANG J. Nuclear receptor HR3 controls locust molt by regulating chitin synthesis and degradation genes of Locusta migratoria[J]. Insect Biochemistry & Molecular Biology, 2018, 92: 1-11. DOI:10.1016/j.ibmb.2017.11.001 |
| [44] |
ZHANG X Y, HE Q H, ZHANG T T, WU H H, ZHANG J Z, MA E B. Characteristics of Halloween genes and RNA interference-mediated functional analysis of LmCYP307a2 in Locusta migratoria[J]. Insect Science, 2022, 29(1): 51-64. DOI:10.1111/1744-7917.12907 |
| [45] |
SHI J F, MU L L, GUO W C, LI G Q. Identification and hormone induction of putative chitin synthase genes and splice variants in Leptinotarsa decemlineata (SAY)[J]. Archives of Insect Biochemistry & Physiology, 2016, 92(4): 242-258. DOI:10.1002/arch.21331 |
| [46] |
ZHAO Z, LI L, CHENG M, JING A D, LIU S N, ZHU S M, DU E X, LI S, LUAN Y X, REN C H. Grainy head signaling regulates epithelium development and ecdysis in Blattella germanica[J]. Insect Science, 2021, 28(2): 485-494. DOI:10.1111/1744-7917.12780 |
| [47] |
PEARSON J C, JUAREZ M T, KIM M, DRIVENES O, MCGINNIS W. Multiple transcription factor codes activate epidermal wound-response genes in Drosophila[J]. Proceedings of the National Academy of Sciences, 2009, 106(47): 20134. DOI:10.1073/pnas.0810219106 |
| [48] |
IBRAHIM G H, SMARTT C T, KILEY L M, CHRISTENSEN B M. Cloning and characterization of a chitin synthase cDNA from the mosquito Aedes aegypti[J]. Insect Biochemistry & Molecular Biology, 2000, 30(12): 1213-1222. DOI:10.1016/s0965-1748(00)00100-4 |
| [49] |
CHEN L, YANG W J, CONG L, XU K K, WANG J J. Molecular cloning, characterization and mRNA expression of a chitin synthase 2 gene from the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae)[J]. International Journal of Molecular Sciences, 2013, 14(8): 17055-17072. DOI:10.3390/ijms140817055 |
| [50] |
刘晓健, 崔淼, 李大琪, 张欢欢, 杨美玲, 张建珍. 飞蝗几丁质合成酶2基因的表达特性、功能及调控[J]. 中国农业科学, 2014, 47(7): 1330-1340. DOI:10.3864/j.issn.0578-1752.2014.07.010 LIU X J, CUI M, LI D Q, ZHANG H H, YANG M L, ZHANG J Z. Expression, function and regulation of chitin synthase 2 gene in Locusta migratoria[J]. Scientia Agricultura Sinica, 2014, 47(7): 1330-1340. DOI:10.3864/j.issn.0578-1752.2014.07.010 |
| [51] |
唐斌, 张露, 熊旭萍, 汪慧娟, 王世贵. 海藻糖代谢及其调控昆虫几丁质合成研究进展[J]. 中国农业科学, 2018, 51(4): 697-707. DOI:10.3864/j.issn.0578-1752.2018.04.009 TANG B, ZHANG L, XIONG X P, WANG H J, WANG S G. Advances in trehalose metabolism and its regulation of insect chitin synthesis[J]. Scientia Agricultura Sinica, 2018, 51(4): 697-707. DOI:10.3864/j.issn.0578-1752.2018.04.009 |
| [52] |
CHEN J, TANG B, CHEN H, YAO Q, HUANG X, CHEN J, ZHANG D, ZHANG W. Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA interference[J]. PLoS One, 2010, 5(4): e10133. DOI:10.1371/journal.pone.0010133 |
| [53] |
TANG B, WEI P, ZHAO L, SHI Z, SHEN Q, YANG M, XIE G, WANG S. Knockdown of five trehalase genes using RNA interference regulates the gene expression of the chitin biosynthesis pathway in Tribolium castaneum[J]. BMC Biotechnology, 2016, 16(1): 67. DOI:10.1186/s12896-016-0297-2 |
| [54] |
ZHAO L, YANG M, SHEN Q, LIU X, SHI Z, WANG S, TANG B. Functional characterization of three trehalase genes regulating the chitin metabolism pathway in rice brown planthopper using RNA interference[J]. Scientific Reports, 2016, 6: 27841. DOI:10.1038/srep27841 |
| [55] |
张露, 朱世城, 郑好, 沈祺达, 王世贵, 唐斌. 褐飞虱海藻糖酶基因在表皮几丁质代谢中的调控作用[J]. 中国农业科学, 2017, 50(6): 10. DOI:10.3864/j.issn.0578-1752.2017.06.006 ZHANG L, ZHU S C, ZHENG H, SHEN Q D, WANG S G, TANG B. Regulatory function of trehalase genes on chitin metabolism in the cuticle of Nilaparvata lugens[J]. Scientia Agricultura Sinica, 2017, 50(6): 10. DOI:10.3864/j.issn.0578-1752.2017.06.006 |
| [56] |
ZHANG L, QIU L Y, YANG H L, WANG H J, ZHOU M, WANG S G, TANG B. Study on the effect of wing bud chitin metabolism and its developmental network genes in the brown planthopper, Nilaparvata lugens, by knockdown of TRE gene[J]. Frontiers in Physiology, 2017, 8: 750. DOI:10.3389/fphys.2017.00750 |
| [57] |
LI Y, CHEN X, WANG S S, PAN B Y, WANG S G, WANG S, TANG B. Evaluation of the expression and function of the TRE2-like and TRE2 genes in ecdysis of Harmonia axyridis[J]. Frontiers in Physiology, 2019, 10: 1371. DOI:10.3389/fphys.2019.01371 |
| [58] |
YU H Z, HUANG Y L, LU Z J, ZHANG Q, SU H N, DU Y M, YI L, ZHONG B L, CHEN C X. Inhibition of trehalase affects the trehalose and chitin metabolism pathways in Diaphorina citri (Hemiptera: Psyllidae)[J]. Insect Science, 2021, 28(3): 718-734. DOI:10.1111/1744-7917.12819 |
| [59] |
LI Y, XU Y, WU S, WANG B, LI Y, LIU Y, WANG J. Validamycin inhibits the synthesis and metabolism of trehalose and chitin in the oriental fruit fly, Bactrocera dorsalis (Hendel)[J]. Insects, 2023, 14(8): 671. DOI:10.3390/insects14080671 |
| [60] |
PAN B Y, LI G Y, WU Y, ZHOU Z S, ZHOU M, LI C. Glucose utilization in the regulation of chitin synthesis in Brown Planthopper[J]. Journal of Insect Science, 2019, 19(5): 3. DOI:10.1093/jisesa/iez081 |
| [61] |
XU CD, LIU YK, QIU LY, WANG SS, PAN BY, LI Y, WANG SG, TANG B. GFAT and PFK genes show contrasting regulation of chitin metabolism in Nilaparvata lugens[J]. Scientific Reports, 2021, 11(1): 5246. DOI:10.1038/s41598-021-84760-2 |
| [62] |
ZOU H, ZHANG B, ZOU C, MA W, ZHANG S, WANG Z, BI B, LI S, GAO J, ZHANG C, ZHANG G, ZHANG J. Knockdown of GFAT disrupts chitin synthesis in Hyphantria cunea larvae[J]. Pesticide Biochemistry and Physiology, 2022, 188: 105245. DOI:10.1016/j.pestbp.2022.105245 |
| [63] |
陈洁, 陈宏鑫, 姚琼, 张文庆. 甜菜夜蛾UAP的克隆、时空表达及RNAi研究[J]. 中国农业科学, 2014, 47(7): 11. CHEN J, CHE H X, YAO Q, ZHANG W Q. Molecular cloning, expression patterns and rnai of UDP-N-acetylglucosamine pyrophosphorylase in Spodoptera exigua[J]. Scientia Agricultura Sinica, 2014, 47(7): 11. |
| [64] |
YANG X B, ZHOU C, GONG M F, YANG H, LONG G Y, JIN D C. Identification and RNAi-based functional analysis of four chitin deacetylase genes in Sogatella furcifera (Hemiptera: Delphacidae)[J]. Journal of Insect Science, 2021, 21(4): 9. DOI:10.1093/jisesa/ieab051 |
| [65] |
YANG X, ZHOU C, LONG G, YANG H, CH EN C, J IN D. Characterization and functional analysis of chitinase family genes involved in nymph-adult transition of Sogatella furcifera[J]. Insect Science, 2021, 28(4): 901-916. DOI:10.1111/1744-7917.12839 |
| [66] |
YANG W J, XU K K, YAN Y, LI C, JIN D C. Role of chitin deacetylase 1 in the molting and metamorphosis of the cigarette beetle Lasioderma serricorne[J]. International Journal of Molecular Sciences, 2020, 21(7): 2449. DOI:10.3390/ijms21072449 |
| [67] |
LIU X Y, WANG S S, ZHONG F, ZHOU M, JIANG X Y, CHENG Y S, DAN Y H, HU G, LI C, TANG B, WU Y. Chitinase (CHI) of Spodoptera frugiperda affects molting development by regulating the metabolism of chitin and trehalose[J]. Frontiers in Physiology, 2022, 13: 1034926. DOI:10.3389/fphys.2022.1034926 |
| [68] |
ZHAO D, LIU X, LIU Z, HANWU, LU X, GUO W. Identification and functional analysis of two potential RNAi targets for chitin degradation in Holotrichia parallela Motschulsky (Insecta Coleoptera)[J]. Pesticide Biochemistry and Physiology, 2022, 188: 105257. DOI:10.1016/j.pestbp.2022.105257 |
| [69] |
XIONG K C, WANG J, LI J H, DENG Y Q, PU P, FAN H, LIU Y H. RNA interference of a trehalose-6-phosphate synthase gene reveals its roles during larval-pupal metamorphosis in Bactrocera minax (Diptera: Tephritidae)[J]. Journal of Insect Physiology, 2016, 91-92: 84-92. DOI:10.1016/j.jinsphys.2016.07.003 |
| [70] |
YANG M M, ZHAO L N, SHEN Q D, XIE G Q, WANG S G, TANG B. Knockdown of two trehalose-6-phosphate synthases severely affects chitin metabolism gene expression in the brown planthopper Nilaparvata lugens[J]. Pest Management Science, 2017, 73(1): 206-216. DOI:10.1002/ps.4287 |
| [71] |
ZHOU M, SHEN Q, WANG S, LI G, WU Y, XU C, TANG B, LI C. Regulatory function of the trehalose-6-phosphate synthase gene TPS3 on chitin metabolism in brown planthopper, Nilaparvata lugens[J]. Insect Molecular Biology, 2022, 31(2): 241-250. DOI:10.1111/imb.12754 |
| [72] |
CHEN Q W, JIN S, ZHANG L, SHEN Q D, WEI P, WEI Z M, WANG S G, TANG B. Regulatory functions of trehalose-6-phosphate synthase in the chitin biosynthesis pathway in Tribolium castaneum (Coleoptera: Tenebrionidae) revealed by RNA interference[J]. Bulletin of Entomological Research, 2018, 108(3): 388-399. DOI:10.1017/S000748531700089X |
| [73] |
WANG G, GOU Y, GUO S, ZHOU J J, LIU C. RNA interference of trem in two color morphs ohalose-6-phosphate synthase and trehalase genes regulates chitin metabolisf Acyrthosiphon pisum Harris[J]. Scientific Reports, 2021, 11(1): 948. DOI:10.1038/s41598-020-80277-2 |
| [74] |
GONG C, YANG Z, HU Y, WU Q, WANG S, GUO Z, ZHANG Y. Silencing of the BtTPS genes by transgenic plant-mediated RNAi to control Bemisia tabaci MED[J]. Pest Management Science, 2022, 78(3): 1128-1137. DOI:10.1002/ps.6727 |
| [75] |
YANG H J, CUI M Y, ZHAO X H, ZHANG C Y, HU Y S, FAN D. Trehalose-6-phosphate synthase regulates chitin synthesis in Mythimna separate[J]. Frontiers in Physiology, 2023, 14: 1109661. DOI:10.3389/fphys.2023.1109661 |
| [76] |
张道伟, 余亚娅, 潘碧莹, 康奎, 曾伯平, 陈静, 唐斌. 白背飞虱海藻糖合成酶基因调控几丁质合成的功能[J]. 中国农业科学, 2019, 52(19): 3357-3366. DOI:10.3864/j.issn.0578-1752.2019.19.007 ZHANG D W, YU Y Y, PAN B Y, KANG K, ZENG B P, CHEN J, TANG B. Regulation function of trehalose-6-phosphate synthase genes on chitin synthesis in Sogatella furcifera[J]. Scientia Agricultura Sinica, 2019, 52(19): 3357-3366. DOI:10.3864/j.issn.0578-1752.2019.19.007 |
| [77] |
PAN B Y, LIU Y K, WU H K, PANG X Q, WANG S G, TANG B, XU C D. Role of phosphoglucomutase in regulating trehalose metabolism in Nilaparvata lugens[J]. Biotechnology, 2020, 10(2): 61. DOI:10.1007/s13205-020-2053-5 |
| [78] |
DING Y J, LI G Y, XU C D, WU Y, ZHOU Z S, WANG S G, LI C. Regulatory functions of Nilaparvata lugens GSK-3 in energy and chitin metabolism[J]. Frontiers in Physiology, 2020, 11: 518876. DOI:10.3389/fphys.2020.518876 |
| [79] |
JOJIMA T, INUI M. Engineering the glycolytic pathway: A potential approach for improvement of biocatalyst performance[J]. Bioengineered, 2015, 6(6): 328-334. DOI:10.1080/21655979.2015.1111493 |
| [80] |
ZHANG S, ZHANG Y, ZOU H, LI X, ZOU H, WANG Z, ZOU C. FDP-Na-induced enhancement of glycolysis impacts larval growth and development and chitin biosynthesis in fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae)[J]. Pesticide Biochemistry and Physiology, 2023, 195: 105560. DOI:10.1016/j.pestbp.2023.105560 |
| [81] |
YANG M, WANG Y, JIANG F, SONG T, WANG H, LIU Q, ZHANG J, ZHANG J, KANG L. miR-71 and miR-263 jointly regulate target genes chitin synthase and chitinase to control Locust molting[J]. PLoS Genetics, 2016, 12(8): e1006257. DOI:10.1371/journal.pgen.1006257 |
| [82] |
CHEN J, LI T, PANG R. miR-2703 regulates the chitin biosynthesis pathway by targeting chitin synthase 1a in Nilaparvata lugens[J]. Insect Molecular Biology, 2020, 29(1): 38-47. DOI:10.1111/imb.12606 |
| [83] |
ZHANG R, LIU W, FU J, ZHANG Z. MicroRNA-989 controls Aedes albopictus pupal-adult transition process by influencing cuticle chitin metabolism in pupae[J]. Parasit Vectors, 2023, 16(1): 397. DOI:10.1186/s13071-023-05976-x |
| [84] |
张一宾. 农药[M]. 北京: 中国物资出版社, 1998. ZHANG Y B. Pesticide[M]. Beijing: China Materials Press, 1998. |
| [85] |
ZHANG J, ZHU K Y. Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron[J]. Insect Biochemistry & Molecular Biology, 2006, 36(9): 712-725. DOI:10.1016/j.ibmb.2006.06.002 |
| [86] |
LU Z J, HUANG Y L, YU H Z, LI N Y, XIE Y X, ZHANG Q, ZENG X D, HU H, HUANG A J, YI L, SU H N. Silencing of the chitin synthase gene is lethal to the Asian Citrus psyllid, Diaphorina citri[J]. International Journal of Molecular Sciences, 2019, 20(15): 3734. DOI:10.3390/ijms20153734 |
| [87] |
ZHANG C, HU W, YU Z, LIU X, WANG J, XIN T, ZOU Z, XIA B. Characterization of chitin synthase A cDNA from Diaphorina citri (Hemiptera: Liviidae) and its response to diflubenzuron[J]. Insects, 2022, 13(8): 728. DOI:10.3390/insects13080728 |
| [88] |
YAO Q, QUAN L F, XU S, DONG Y Z, LI WJ, CHEN B X. Effect of diflubenzuron on the chitin biosynthesis pathway in Conopomorpha sinensis eggs[J]. Insect Science, 2021, 28(4): 1061-1075. DOI:10.1111/1744-7917.12848 |
| [89] |
刘晓健, 杨美玲, 张建琴, 马恩波, 张建珍. 氟虫脲对东亚飞蝗和中华稻蝗几丁质合成酶基因表达的影响[J]. 昆虫学报, 2010, 53(9): 1039-1044. DOI:10.16380/j.kcxb.2010.09.014 LIU X J, YANG M L, ZHANG J Q, MA E B, ZHANG J Z. Effects of flufenuron on the expression of chitin synthase gene in Locusta migratoria (Meyen) (Orthoptera: Acrididae) and Oxyachinensis (Thunberg) (Orthoptera: Oedipodidae)[J]. Acta Entomologica Sinica, 2010, 53(9): 1039-1044. DOI:10.16380/j.kcxb.2010.09.014 |
| [90] |
CHANG C L, GEIB S, CHO I K, LI Q X, STANLEY D. Dietary lufenuron reduces egg hatch and influences protein expression in the fruit fly Bactrocera latifrons (Hendel)[J]. Archives of Insect Biochemistry & Physiology, 2014, 86(4): 193-208. DOI:10.1002/arch.21169 |
| [91] |
陈金鹏, 谢佳, 赵柯, 胡展, 张羲, 南晓慧, 孙然锋. 新型2, 6-二氟苯甲酰脲类化合物对草地贪夜蛾的生物活性及其几丁质合成的影响[J]. 农药学学报, 2023, 25(4): 798-807. DOI:10.16801/j.issn.1008-7303.2023.0046 CHEN J, XIE J, ZHAO K, HU Z, ZHANG X, NAN X, SUN R. Effects of a novel 2, 6-difluorobenzoylurea compound on the bioactivity and chitin synthesis of Spodoptera frugiperda[J]. Chinese Journal of Pesticide Science, 2023, 25(4): 798-807. DOI:10.16801/j.issn.1008-7303.2023.0046 |
| [92] |
台术蕾, 马龙, 张春妮. 棉铃虫几丁质合成酶1基因的时空表达分析及氟啶脲对其表达的影响[J]. 环境昆虫学报, 2020, 42(1): 137-146. DOI:10.3969/j.issn.1674-0858.2020.01.17 TAI S L, MA L, ZHANG C N. Spatio-temporal expression analysis of chitin synthase 1 gene in Helicoverpa armigera and its responses to chlorfluazuron[J]. Journal of Environmental Entomology, 2020, 42(1): 137-146. DOI:10.3969/j.issn.1674-0858.2020.01.17 |
| [93] |
COHEN E. Chitin biochemistry: Synthesis and inhibition[J]. Annual Review of Entomology, 1987, 32: 71-93. DOI:10.1146/annurev.en.32.010187.000443 |
| [94] |
ULLAH F, GUL H, YOUSAF H K, XIU W, QIAN D, GAO X, TARIQ K, HAN P, DESNEUX N, SONG D. Impact of low lethal concentrations of buprofezin on biological traits and expression profile of chitin synthase 1 gene (CHS1) in melon aphid, Aphis gossypii[J]. Scientific Reports, 2020, 10(1): 18158. DOI:10.1038/s41598-019-48199-w |
| [95] |
ZHAO L, WEI P, GUO H, WANG S, TANG B. Suppressing the expression of a forkhead transcription factor disrupts the chitin biosynthesis pathway in Spodoptera exigua[J]. Archives of Insect Biochemistry and Physiology, 2014, 86(1): 4-18. DOI:10.1002/arch.21145 |
| [96] |
YUE X, LIANG Y, WEI Z, LYU J, CAI Y, FAN X, ZHANG W, CHEN J. Genome-wide in vitro and in vivo RNAi screens reveal Fer3 to be an important regulator of kkv transcription in Drosophila[J]. Insect Science, 2022, 29(3): 614-630. DOI:10.1111/1744-7917.12954 |
| [97] |
XU G, ZHANG J, LYU H, SONG Q, FENG Q, XIANG H, ZHENG S. DNA methylation mediates BmDeaf1-regulated tissue-and stage-specific expression of BmCHSA-2b in the silkworm, Bombyx mori[J]. Epigenetics & Chromatin, 2018, 11(1): 32. DOI:10.1186/s13072-018-0202-4 |
| [98] |
MCHALE C M, ZHANG L, LAN Q, VERMEULEN R, LI G, HUBBARD A E, PORTER K E, THOMAS R, PORTIER C J, SHEN M, RAPPAPORT S M, YIN S, SMITH M T, ROTHMAN N. Global gene expression profiling of a population exposed to a range of benzene levels[J]. Environmental Health Perspectives, 2011, 119(5): 628-634. DOI:10.1289/ehp.1002546 |
| [99] |
PAJARO-CASTRO N, CABALLERO-GALLARDO K, OLIVEROVERBEL J. Toxicity of naphthalene and benzene on Tribollium castaneum Herbst[J]. International Journal of Environmental Research and Public Health, 2017, 14(6): 667. DOI:10.3390/ijerph14060667 |
| [100] |
FRUTTERO L L, MOYETTA N R, KRUG M S, BROLL V, GRAHL M V, REAL-GUERRA R, STANISÇUASKI F, CARLINI C R. Jaburetox affects gene expression and enzyme activities in Rhodnius prolixus, a Chagas' disease vector[J]. Acta Tropica, 2017, 168: 54-63. DOI:10.1016/j.actatropica.2017.01.009 |
| [101] |
陈焕瑜, 胡珍娣, 冯夏, 李振宇, 张德雍. 粤中地区小菜蛾对啶虫隆的抗性监测及治理对策[J]. 广东农业科学, 2010, 37(9): 30-31. DOI:10.16768/j.issn.1004-874X.2010.09.025 CHEN H Y, HU Z D, FENG X, LI Z Y, ZHANG D Y. Resistance monitoring and control measures of Plutella xylostella to pyridon in central Guangdong Province[J]. Guangdong Agricultural Sciences, 2010, 37(9): 30-31. DOI:10.16768/j.issn.1004-874X.2010.09.025 |
| [102] |
朱本全, 杜馨. 昆虫海藻糖酶及其抑制剂研究进展[J]. 生物化工, 2019, 5(6): 3. DOI:10.3969/j.issn.2096-0387.2019.06.046 ZHU B Q, DU X. Insect trehalase and its inhibitors[J]. Biological Chemical Engineering, 2019, 5(6): 3. DOI:10.3969/j.issn.2096-0387.2019.06.046 |
| [103] |
BOIJA A, KLEIN I A, SABARI B R, DALL'AGNESE A, COFFEY E L, ZAMUDIO A V, LI C H, SHRINIVAS K, MANTEIGA J C, HANNETT N M, ABRAHAM B J, AFEYAN L K, GUO Y E, RIMEL J K, FANT C B, SCHUIJERS J, LEE T I, TAATJES D J, YOUNG R A. Transcription factors activate genes through the phase-separation capacity of their activation domains[J]. Cell, 2018, 175(7): 1842-1855. DOI:10.1016/j.cell.2018.10.042 |
| [104] |
HENNINGER J E, OKSUZ O, SHRINIVAS K, SAGI I, LEROY G, ZHENG M M, ANDREWS J O, ZAMUDIO A V, LAZARIS C, HANNETT N M, LEE T I, SHARP P A, CISSÉ I I, CHAKRABORTY A K, YOUNG R A. RNA-mediated feedback control of transcriptional condensates[J]. Cell, 2021, 184(1): 207-225. DOI:10.1016/j.cell.2020.11.030 |
(责任编辑 陈丽娥)
 2024, Vol. 51
2024, Vol. 51