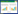文章信息
基金项目
- 国家自然科学基金(31901441);广东省农业科学院科技创新战略建设专项资金(高水平农科院建设)项目(R2021PY-QF001);广东省水稻育种新技术重点实验室项目(2023B1212060042)
作者简介
- 韩明珍(1997—),女,在读硕士生,研究方向为水稻逆境生理,E-mail:hmz109927@163.com.
通讯作者
- 周玲艳(1972—),女,博士,教授,研究方向为植物生物技术,E-mail:lingyanzh@163.com; 马雅美(1991—),女,博士,副研究员,研究方向为水稻逆境生理,E-mail:mayamei@gdaas.cn.
文章历史
- 收稿日期:2024-03-06
2. 广东省农业科学院水稻研究所/广东省水稻育种新技术重点实验室/广东省水稻工程实验室/农业农村部华南优质稻遗传育种实验室(部省共建),广东 广州 510640;
3. 广东海洋大学滨海农业学院,广东 湛江 524088;
4. 华南农业大学生命科学学院,广东 广州 510640
2. Rice Research Institute, Guangdong Academy of Agricultural Sciences / Guangdong Key Laboratory of New Technology in Rice Breeding / Guangdong Rice Engineering Laboratory / Key Laboratory of Genetics and Breeding of High Quality Rice in Southern China (Co-construction by Ministry and Province), Ministry of Agriculture and Rural Aff airs, Guangzhou 510640, China;
3. College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang 524088, China;
4. College of Life Sciences, South China Agricultural University, Guangzhou 510640, China
水稻(Oryza sativa L.)是世界主要粮食作物之一,也是全球超过半数人口的食物来源。水稻高产稳产对保证世界粮食安全、社会稳定和人民生活水平具有极其重要的作用。为了满足人口增长的需求,预计到2050年,粮食产量需要翻一番[1]。由于水稻生长对水分要求较高,随着全球气候条件变化(如变暖)及农业领域内水资源稀缺状况的日益严峻,干旱已成为限制水稻产量的主要因素之一[2]。研究表明,水稻抗旱性属于复杂的数量性状[3]。随着测序技术和基因组学的快速发展,目前已经发现了大量参与干旱胁迫应答的基因。根据抗旱相关基因功能的差异,其主要分为3类:(1)信号转导基因,如蛋白激酶;(2)转录因子基因,如bZIP、MYB/MYC、WRKY、NAC、DREB等编码基因;(3)功能蛋白编码基因,如与渗透调节物质合成和氧化还原系统相关的编码基因[4-5]。这些基因通过参与干旱信号的应答、信号传导及基因表达调控等过程完成干旱适应性反应。其中,转录因子在水稻干旱胁迫响应基因的表达调控中起着关键作用。
植物细胞质膜上存在一系列特定的受体蛋白,它们扮演着感知干旱信号的关键角色。当植物受到干旱胁迫时,这些受体蛋白如G蛋白偶联受体(Gprotein-coupled receptors,GPCRs)、受体类似激酶(Receptor-like kinases,RLKs)、组氨酸磷酸激酶(Histidine kinases,HKs)、脱落酸(Abscisic acid,ABA)受体及钙感应受体(Calcium-sensing receptor,CAS)等,能够灵敏地捕获并响应外界的干旱胁迫信号。胁迫信号会通过磷脂信号系统启动一系列复杂的生化反应,进而生成多种第二信使分子,如Ca2+、活性氧(Reactive oxygen species,ROS)、三磷酸肌醇(Inositol 1, 4, 5-triphosphate,IP3)和二酰甘油(Diacylglycerol,DAG)等,其在细胞内起着传递和放大信号的作用以激活下游相应的信号通路。转录因子是指能够特异性结合某基因启动子区域顺式作用元件的蛋白质。作为主要调控因子之一,转录因子可以将胁迫信号转化为最终的细胞反应。受激发的转录因子能够与下游调控基因启动子的顺式作用元件如G-box、MYBRS/MYCRS、NACR、DRE、W-box等结合,激活或抑制该基因的表达,进而引起植物渗透调节及抗氧化系统的改变,最终使植物适应干旱胁迫逆境或增强植物自身的抗旱能力[6](图 1)。目前,植物中参与干旱胁迫反应的转录因子主要有bZIP、MYB、NAC、DREB、WRKY等。本文对水稻干旱胁迫响应中涉及的转录因子的调控作用进行综述。
1 bZIP型转录因子
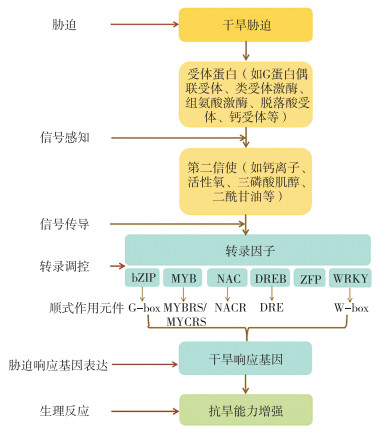
碱性亮氨酸拉链(Basic leucine zipper,bZIP)转录因子含有保守的、由60~80个氨基酸组成的bZIP结构域。bZIP结构域由1个碱性(Basic)区域和1个富含亮氨酸的拉链状(Leucine zipper,LZ)结构组成[7-8]。水稻基因组内含有89个bZIP型转录因子,不均匀地分布在水稻12条染色体上[9]。另有报道指出,33个bZIP型转录因子的表达量受干旱胁迫影响,其中24个可被诱导表达,9个被抑制表达[9]。目前,OsbZIP03/OsHBP1b、OsbZIP05、OsABI5/OsbZIP10、OsbZIP12、OsbZIP16、OsbZIP20、OsbZIP23、OsbZIP33、OsbZIP40、OsbZIP45、OsbZIP46、OsbZIP52、OsbZIP60、OsbZIP62、OsbZIP66、OsbZIP71、OsbZIP72、OsbZIP86在水稻干旱胁迫应答中的功能已被深入探究与解析[10-29](表 1)。
OsbZIP23是ABA-依赖途径中的核心调控因子,它的表达受干旱、ABA、低温等强烈诱导。过量表达OsbZIP23可导致水稻对于ABA处理更加敏感,并可提高水稻植株的抗旱性和耐盐性,反之,则osbzip23突变体的抗旱性和耐盐性显著降低[16]。RNA-seq和ChIP-Seq的数据表明,OsbZIP23通过结合G-box(CACGTG)顺式作用元件直接调控众多已知在胁迫响应、激素信号等过程中发挥功能的基因,如OsPP2C49(Clade A protein phosphatase 2C)等[17]。本团队研究发现,水稻14-3-3蛋白OsGF14f(G-box factor 14-3-3 homologs)可以通过影响OsbZIP23对下游靶标基因如OsLEA3-2(Late embryogenesis abundant protein 3-2)的转录调控活性从而共同调控水稻的抗旱性,同时OsGF14f又是OsbZIP23的下游靶标,从而形成正反馈调控[18]。OsbZIP46也是ABA-依赖途径中重要的调节因子,然而与OsbZIP23不同,OsbZIP46的D结构域对其自身转录激活功能具有抑制作用,因此只有过量表达OsbZIP46CA1(去除D结构域的OsbZIP46)才可以显著提高水稻抗旱性。RNA-seq结果显示,OsbZIP46和OsbZIP23调控的下游基因有明显差异,表明OsbZIP23和OsbZIP46可能通过不同的调控途径参与干旱胁迫应答[22]。OsbZIP71的表达可以被干旱、PEG和ABA强烈诱导,其在ABA介导的干旱胁迫耐受性过程中发挥着重要作用。研究表明,OsbZIP71可通过G-box直接与OsNHX1(Vacuolar Na+/H+ antiporter gene)和COR413-TM1(Cold acclimation protein 413-TM1)这两个逆境胁迫相关基因的启动子结合并激活它们的表达,从而提高水稻的抗旱和抗盐性[27]。Gao等[29]研究发现,OsbZIP86可以结合ABA合成相关基因OsNCED3(9-cis-epoxycarotenoid dioxygenase)的启动子从而正向调控水稻的抗旱性;miR2105可靶向剪切OsbZIP86的mRNA,以实现负向调控水稻抗旱性;蛋白激酶OsSAPK10(Stress/ABA-activated protein kinase)可以磷酸化OsbZIP86,并增强OsbZIP86对于OsNCED3的转录激活活性,以上结果暗示了miR2105-(OsSAPK10)-OsbZIP86-OsNCED3模块在水稻干旱胁迫应答中发挥着重要作用。类似地,过量表达OsbZIP20显著提高了水稻对盐和干旱胁迫的耐受性,且OsSAPK10与OsbZIP20互作可将其磷酸化,磷酸化的OsbZIP20增强了其与OsNHX1启动子中ABRE(ABA responsive element binding protein)元件的结合能力,并诱导OsNHX1表达,最终赋予水稻植株更高的抗旱能力[15]。
与以上正向调控水稻抗旱性的OsbZIP转录因子不同,水稻中还存在一些负向调控抗旱性的OsbZIP成员。比如,在20%PEG模拟干旱胁迫的条件下,相比于野生型,OsbZIP05-RNAi转基因水稻幼苗的脯氨酸及叶绿素含量升高、丙二醛含量和失水速率降低、抗旱性增强[11]。过量表达OsbZIP52会导致水稻抗旱性显著下降,同时响应干旱胁迫的相关基因如OsLEA3、OsTPP1(Trehalose-6-phosphate phosphatase 1)、Rab25(Responsive to ABA 25)、gp1 precursor、β-gal、OsGELP63(GDSL esterase/lipase 63)、LOC_Os05g39250的表达量下降[23],表明OsbZIP05和OsbZIP52是负调控水稻抗旱性的蛋白因子。
2 MYB转录因子MYB(v-myb avain myeloblastosis viral oncogene homolog)转录因子家族是植物体中成员较多的转录因子家族之一,因其含有高度保守的MYB结构域而得名。一般来说,MYB结构域由1~4个不完全重复序列组成,每个重复序列具有约52个氨基酸残基,这些氨基酸呈螺旋-旋转-螺旋的构象插入双链DNA主沟中[30]。根据MYB结构域的数量,该家族可分为4个亚群:1R、R2R3、3R和4R[31],其中R2R3亚群的成员在植物的生长发育、生理代谢及逆境响应过程中起着关键作用。目前,在水稻基因组中累计发现了239个MYB基因[32],这些基因已被大量报道和证实能直接或者间接参与水稻逆境胁迫响应。目前,OsMYB2、OsMYB6、OsMYB26、OsMYB48-1、OsMYB60、OsFLP、OsMYB1R1、OsMYB3R-2等MYB基因参与调控水稻抗旱性的功能和分子机制被陆续报道[25, 33-39](表 1)。
OsMYB2受干旱诱导,过量表达OsMYB2可以通过提高过氧化物酶(Peroxidase,POD)、超氧化物歧化酶(Superoxide dismutase,SOD)等抗氧化酶的活性,减少H2O2和丙二醛的含量,从而使水稻获得更强的抗旱性[33]。OsMYB48-1的表达受PEG、ABA、H2O2和干旱胁迫的诱导,且过量表达OsMYB48-1能增强水稻对干旱和NaCl的耐受性[35]。相似地,OsMYB6在面临干旱和NaCl胁迫时也会被诱导表达,过量表达OsMYB6可以赋予水稻更强的抗旱和耐盐性,且不影响其正常生长发育[34]。另外,干旱也可以显著诱导OsMYB60的表达,相比于野生型,过量表达OsMYB60可以显著提高水稻的抗旱性。研究表明,OsMYB60可以通过MYB结合位点(T/AAACCA或者T/CAACTA/G)直接结合蜡质合成相关基因OsGL1-4/OsCER1(Glossy1-homologous gene)的启动子并激活其表达,从而提高水稻的抗旱能力[36]。近期研究发现,R2R3亚群MYB转录因子基因OsFLP的表达受干旱、NaCl、ABA等诱导。高表达OsFLP可以促进气孔关闭,使植株的抗旱性提高。此外,酵母单杂实验结果表明OsFLP可以结合到NAC转录因子基因OsNAC1和OsNAC6的启动子区域,暗示OsFLP可能通过调控OsNAC1和OsNAC6的转录从而共同响应干旱胁迫[37]。
与上述正向调控水稻抗旱性的MYB转录因子不同,OsMYB1R1、OsMYB26等MYB基因家族成员则负向调控水稻抗旱性。干旱处理抑制OsMYB1R1基因的转录。正常生长条件下,OsMYB1R1-RNAi、OE植株的生长与野生型相比无明显差异,但是在干旱条件下,相比于野生型,OsMYB1R1-RNAi植株体内的脯氨酸含量较高,而丙二醛含量和电导率较低,最终提高了OsMYB1R1-RNAi植株的存活率[38]。此外,OsMYB26可以通过MYB结合位点结合LEA3基因的启动子从而负向调控水稻抗旱性,且干旱可诱导OsMFT1(Mother of FT and TFL1)和OsbZIP66的表达,抑制OsFTIP1(FT-interacting protein1)和OsMYB26的表达[26]。由于OsFTIP1含量降低,导致进入细胞核的OsMFT1蛋白增多,在细胞核内,OsMFT1分别与OsMYB26和OsbZIP66互作,一方面减弱OsMYB26对LEA3等基因的抑制作用,另一方面增强OsbZIP66对Rab21(Responsive to ABA 21)等基因的激活作用,从而完成干旱应答[26]。
3 NAC转录因子NAC(NAM、ATAFs和CUC)转录因子的命名源自于矮牵牛(Petunia hybrida)的NAM(No apical meristem)基因、拟南芥(Arabidopsis thaliana)的ATAF1/2基因和CUC2(Cup - shaped cotyledon)基因的首字母。该类转录因子广泛分布于陆生植物中,参与植物次生壁生长、侧根生长、叶片衰老和次生代谢等生长发育过程,以及干旱、高盐、病原菌感染等逆境胁迫应答[40-41]。NAC蛋白具有高度保守的N端DNA结构域和可变的C端转录调控结构域[42]。目前,在水稻基因组中已鉴定出超过170个NAC基因[43],这些基因在逆境胁迫响应(如干旱、低温、高盐等)过程中发挥重要作用。已有多个NAC成员,诸如SNAC1/OsNAC19/OsNAC9、SNAC2/OsNAC6、SNAC3/ONAC003、OsNAC2/OsTIL1/
OMTN2/OsORE1、OsNAC5、OsNAC10、OsNAC14、OsNAC17、ONAC022、OsNAC45/ONAC045、OsNAC58/OsNAP、ONAC066、ONAC092、ONAC095、OsDSR42等被发现参与干旱胁迫应答[44-63](表 1)。
SNAC1(OsNAC19/OsNAC9)的表达受盐、干旱、低温和ABA处理的诱导。在干旱胁迫下,SNAC1主要在保卫细胞中被诱导表达[44]。过量表达SNAC1可显著增强转基因水稻植株在营养生长期的耐旱性和耐盐性及穗期的耐旱性[44]。目前,已鉴定到多个调控水稻逆境胁迫响应的SNAC1下游靶基因。比如,SNAC1可直接与OsSRO1c(Similar to RCD1 gene)的启动子结合并激活其表达。激活的OsSRO1c可以促进气孔关闭、减少H2O2积累,从而正向调控水稻抗旱性[45]。此外,SNAC1也可与OsPP18(Protein Phosphatase18)的启动子结合并调控其表达。OsPP18通过不依赖ABA的ROS清除途径正向调控水稻的抗旱性和抗氧化胁迫[46]。SNAC1的功能也受到其上游调控因子的精确调控,比如IPA1(Ideal plant architectutre1)可直接与SNAC1的启动子结合并促进SNAC1的表达,提高水稻体内的抗氧化还原系统活性,最终赋予水稻植株更强的抗旱能力[47]。OsNAC2(OsTIL1/OsMTN2/OsORE1)的表达受到ABA和干旱、高盐处理的强烈诱导。OsNAC2过量表达植株的抗旱性降低,而OsNAC2-RNAi植株在营养生长期和生殖生长期的抗旱性均得到增强,表明OsNAC2是干旱胁迫响应的负调控因子[50]。OsNAC5的表达受到干旱、冷、高盐等非生物胁迫及ABA和茉莉酸甲酯的诱导。在水稻根部特异性过量表达OsNAC5可以使根部增粗,从而提高水稻的抗旱能力和产量[51]。进一步研究表明,OsNAC5与OsNAC6和SNAC1互作,且OsNAC5可以与OsLEA3、OsCCR10(Cinnamoyl-CoA reducatse)的启动子结合,并激活它们的表达[52-53]。OsCCR10编码一个肉桂酰辅酶A还原酶,参与H-和G-木质素的生物合成,可以通过调节木质素积累的含量来调控水稻的抗旱性[53]。SNAC2(ONAC6/OsNAC6)可以通过调控根部发育来增强抗旱性。经过多年干旱田间试验表明,OsNAC6根特异性过表达转基因水稻植株受干旱胁迫的影响小于非转基因对照植株。RNA-seq结果显示,OsNAC6可以上调参与膜修饰、烟酰胺生物合成等基因的表达。其中,过量表达烟酰胺生物合成酶基因可以促进金属螯合剂的积累,从而使植株的耐旱性提高[48]。OsNAC14主要在减数分裂期表达,且干旱、高盐、低温、ABA处理均可诱导其表达。田间干旱试验表明,过表达OsNAC14转基因水稻品系的穗数和灌浆率均高于非转基因水稻植株,后续研究证实OsNAC14可与DNA修复系统中同源重组关键组分OsRAD51A1的启动子直接相互作用,表明OsNAC14可能通过DNA损伤修复途径来参与调控水稻抗旱性[55]。在干旱胁迫条件下,过表达ONAC066提高了转基因水稻对干旱和氧化胁迫的耐受性,增加了其对ABA的敏感性。相对地,ONAC066-RNAi植株对干旱和氧化胁迫的耐受性降低。此外,ONAC066可以直接与OsDREB2A的启动子结合,并激活OsDREB2A的转录[60]。
4 DREB转录因子DREB(Dehydration responsive element binding protein)又称为C重复序列结合因子(C-repeat binding factor,CBF)蛋白,属于AP2/ERF(APETALA2/Ethylene response element binding factors)转录因子家族的一个亚家族[64]。研究表明,DREB转录因子通过与启动子区域具有DRE/CRT顺式作用元件的逆境胁迫响应基因特异性结合来调控这类基因的表达,从而参与植物逆境胁迫响应过程[64]。目前,水稻基因组中已经鉴定出57个DREB转录因子[65]。根据其序列结构的不同可以分划分为6个亚组:A1、A2、A3、A4、A5和A6,其中A1/OsDREB1主要参与低温胁迫应答,A2/OsDREB2则在干旱胁迫应答过程中发挥作用,其他亚组研究尚较有限(表 1)。
过量表达A1/OsDREB1型转录因子(OsDREB1A、OsDREB1B、OsDREB1F、OsDREB1G)均可以提高转基因水稻植株的抗旱性,然而与野生型相比,过表达OsDREB1E的水稻植株其抗旱能力未产生显著性变化[66-67]。类似地,A2/OsDREB2型转录因子也参与调控干旱、高盐和高温等胁迫的应答。OsDREB2A的表达受干旱和ABA的诱导,并且正向调控水稻抗旱性[68]。A2/OsDREB2型转录因子OsDREB2B存在功能性转录本(OsDREB2B2)和无功能性转录本(OsDREB2B1),其中功能性转录本OsDREB2B2受干旱、高温、低温和高盐等胁迫诱导表达,过量表达OsDREB2B2可以显著提高水稻的抗旱性[69]。OsDRAP1是新鉴定的A2/DREB2型转录因子,该基因通过调控水分平衡、氧化还原平衡及维管束发育等正向调控水稻抗旱性。进一步研究表明,OsDRAP1通过激活OsCBSX3(a Cystathionine beta-synthase domain containing protein)等干旱响应基因的表达,从而共同调控水稻的抗旱性[65]。此外,Ke等[70]发现A6型DREB基因OsDREB6能被干旱、高盐和低温诱导表达,其可通过提高水稻植株体内的可溶性糖、脯氨酸和过氧化氢酶(Catalase,CAT)的含量并降低其植株体内的丙二醛含量来增强水稻的抗旱性。
5 WRKY转录因子WRKY是植物中特有的一类转录因子家族,含有保守的WRKYGQK(色氨酸-精氨酸-赖氨酸-酪氨酸-甘氨酸-谷氨酰胺-赖氨酸)结构域。WRKY通过识别W-box序列和一些含有TGAC核心结构的类似W-box序列,并与之特异性结合,最终调控下游靶基因的表达[71]。水稻基因组中含有103个编码WRKY转录因子的基因,其中有19个WRKY基因的表达受干旱胁迫影响[72]。迄今,科学家们陆续发现OsWRKY08、OsWRKY11、OsWRKY30、OsWRKY47、OsWRKY76、OsWRKY97等转录因子正向调控水稻的抗旱性,OsWRKY5、OsWRKY45、OsWRKY67等则负向调控水稻的抗旱性[73-81](表 1)。
OsWRKY11被证实可正向调控水稻白叶枯病抗性和抗旱性。研究表明,过表达OsWRKY11的转基因植株表现出更强抵御白叶枯病菌和干旱胁迫的能力,因为OsWRKY11能够与干旱诱导响应基因Rab21的启动子结合并激活其表达[74]。OsWRKY30受干旱和ABA诱导表达,其蛋白可以与众多丝裂原活化蛋白激酶互作,比如OsMPK3(Mitogen-activated protein kinase)、OsMPK4、OsMPK7、OsMPK14、OsMPK20-4和OsMPK20-5,且OsMPK3、OsMPK7和OsMPK14均可磷酸化OsWRKY30。过表达OsWRKY30能够增强转基因水稻植株对干旱的耐受性,然而过表达OsWRKY30AA(所有的丝氨酸替换成丙氨酸)的转基因植株其抗旱性相比于野生型无显著差异,表明OsWRKY30的磷酸化水平对其正常功能的发挥有重要影响[75]。OsWRKY47属于Ⅱ型WRKY家族,oswrky47突变体植株产量降低且对干旱的敏感性增强,与之相反,过表达植株的抗旱能力增强;生物信息学分析显示,OsWRKY47可以影响钙调素结合蛋白基因CaMBP及半胱氨酸分泌蛋白基因CRRSP(Cysteine-rich Receptor-like Secreted Protein)的表达[76]。OsWRKY76的表达受PEG、干旱等诱导,酵母双杂和BiFC实验表明,OsWRKY76可与OsJAZ12(Jasmonate ZIM-domain protein)互作,从而削弱了OsJAZ12与OsbHLH148之间的互作,进一步研究发现,OsWRKY76能够部分解除OsJAZ12对OsbHLH148转录激活活性的抑制作用,同时,OsWRKY76和OsbHLH148可以与OsDREB1E启动子结合并激活其表达,共同调控水稻抗旱性[77]。吕苗苗[78]通过对水稻OsWRKY97过表达植株进行多种胁迫处理,发现相比于对照组‘日本晴’,OsWRKY97过表达植株具有更强的抗旱性且对ABA处理更加敏感。
此外,部分WRKY转录因子也可负向调控水稻的抗旱性。比如,OsWRKY5主要在幼苗和穗期的叶片中表达,干旱胁迫、NaCl、甘露醇和ABA处理均会抑制该基因的表达,其突变体的抗旱性增强、对ABA的敏感性提高,从而促进了ABA依赖性的气孔关闭。双荧光素酶、酵母单杂交和染色质免疫沉淀实验表明,OsWRKY5通过直接结合OsMYB2启动子区域的W-box序列来抑制OsMYB2的表达,从而下调ABA信号通路中OsMYB2下游基因的表达[79]。在OsWRKY45位点上有2个等位基因OsWRKY45-1和OsWRKY45-2,它们的编码产物之间仅有10个氨基酸的差异。OsWRKY45-1可负向调控水稻ABA信号,而OsWRKY45-2正向调控ABA信号,但它们均负向调控水稻的抗旱性[80]。另外,OsWRKY67的表达受低温、干旱、高盐和ABA等处理的影响,沉默OsWRKY67基因可显著增强水稻的耐旱性,进一步研究发现,该基因可调控逆境胁迫相关基因OsDREB1A、OsDREB1B、OsDREB2A、OsDREB2B,OsRaB16A(Responsive to ABA 16A)、OsLIP9(Li-pase 9)和OsLEA3等的表达及ABA合成相关基因OsNCED3和OsNCED4等的表达[81]。
6 锌指蛋白转录因子锌指蛋白(Zinc-finger protein,ZFP)因其具有指状结构且能与锌结合而得名。根据锌指蛋白在指状二级结构中与Zn2+结合的半胱氨酸和组氨酸残基的数量和顺序不同,可将锌指蛋白分为9大类[82]。锌指蛋白已被证实参与植物种子萌发及成熟、花器官及配子发育、细胞凋亡等生长发育过程及调控低温、高盐、干旱、白叶枯病等逆境胁迫过程。目前,在水稻中已鉴定出与干旱相关的锌指蛋白有OsZFP252(RZF71)、DST、OsCOIN、OsSAP1、OsiSAP7、OsiSAP8、OsC3H47、OsMSR15、OsBIRF1、OsRHP1、OsDRZ1、OsZFP37和OsC3H等[83-94](表 1)。
DST是水稻中发现的一个新型锌指转录因子,该转录因子通过影响ROS的积累和调节气孔开度来负向调控水稻的抗旱性[84]。后续的研究表明,DST能够与自身物理互作,同时可与DCA1(DST Co-activator 1)形成异源四聚体,该复合物可以直接调控编码H2O2清除因子Prx24基因的表达来影响植株干旱和盐胁迫耐受性[95]。Leaf Panicle 2(LP2)编码细胞膜类受体蛋白激酶,LP2的表达受DST的直接调控,过量表达LP2会导致水稻植株失水速率加快和气孔关闭受阻,从而降低了植株对干旱胁迫的耐受性[96]。
OsDRZ1属于C2H2型锌指蛋白,过量表达OsDRZ1可通过增加脯氨酸的含量、减少ROS的积及并增强抗氧化还原酶的活性来提高水稻的抗旱性。另外,类萌发素蛋白基因OsGLP1(germin-like protein1)可能是OsDRZ1的下游靶基因[93]。OsCOIN是一个环型锌指蛋白,其表达受到干旱胁迫的强烈诱导,过量表达OsCOIN可上调OsP5CS(△ 1-pyrroline-5-carboxylate synthetase)的表达和提高脯氨酸含量等,进而增强水稻对干旱胁迫的耐受性[85]。OsSAP1(OsiSAP1)是一种A20/AN1锌指蛋白转录因子基因,它受冷、干旱、盐、淹涝等多种不同胁迫的诱导,可通过影响转录因子基因、膜转运蛋白、生长发育和参与代谢等相关基因的表达来正向调控水稻的抗旱性[86]。进一步研究发现,OsSAP1通过与转氨酶OsAMTR1(Aminotransferase)及病程相关蛋白OsSCP/OsPR1a(Pathogenesis-related protein 1a)互作来共同调控胁迫应答信号[97]。OsMSR15编码C2H2型锌指蛋白,酵母单杂交实验表明,OsMSR15具有转录激活活性。在拟南芥中异源表达OsMSR15能显著增强转基因植株的抗旱能力及对ABA的敏感性[90]。Zeng等[92]在水稻中鉴定到一个RING-H2锌指蛋白基因OsRHP1,相比于野生型,过表达OsRHP1的水稻植株体内含有更多的ABA,从而赋予植株更高的抗旱性和耐盐性。此外,过量表达的OsZFP37和OsC3H可以与Osr40C1(Euonymus europaeus lectin)的启动子结合并激活其表达,从而进一步提高水稻的抗旱性[94]。
7 其他转录因子除了以上主要的转录因子家族,参与水稻干旱胁迫响应的转录因子还有SPL蛋白(Squamosa promoter binding protein-like family)、bHLH蛋白(Basic helix-loop-helix proteins)、同源异形结构域转录因子家族(Homeodomain leucine zipper proteins,HD-Zip)、MADS转录因子、转录因子热应激蛋白(Heat shock transcrirtional factors,HSF)和GRAS转录因子。近年来,科学家们陆续对这些转录因子在水稻干旱胁迫应答中的功能和机制进行了研究(表 1)。
7.1 SPL转录因子SPL转录因子序列中含有一个SBP结构域(Squamosa promoter-binding protein domain),该转录因子家族广泛参与调控植物胚胎发育、营养生长、花发育、育性发育及次生代谢等过程,然而其参与的表达调控机制仍需要进一步明晰[98-99]。水稻中共发现19个OsSPL转录因子[100],其中OsSPL4主要在完全展开的叶及根中表达。经过干旱处理后,相比于过表达植株和野生型植株,osspl4突变体植株在复水后的存活率显著提高,同时其内部的SOD、POD、CAT等酶活性也明显高于野生型和OsSPL4-OE过表达植株,暗示了OsSPL4负向调控水稻的抗旱性[101]。Li等[102]发现OsSPL10通过控制ROS产生和气孔运动在调控水稻抗旱性中发挥了重要作用。通过对OsSPL10的单倍型和等位基因频率进行分析,得出OsSPL10Hap1等位基因主要存在于大多数旱稻品种中,而OsSPL10Hap2等位基因主要存在于水稻品种中。且在干旱胁迫下,具有OsSPL10Hap1等位基因的品种中,OsSPL10及其下游靶基因OsNAC2的表达量较低,从而降低了OsAP37(Transcription factor with the AP2 domain)的表达并增加了OsCOX11(Cytochrome c oxidase assembly protein)的表达以防止ROS积累和细胞程序性死亡。类似地,敲除OsSPL10可以诱导气孔快速关闭并防止水分流失,从而提高水稻的抗旱性。相反,在含有OsSPL10Hap2等位基因的品种中,OsSPL10的高表达促进了OsNAC2的表达,导致ROS过度积累和细胞程序性死亡,最终造成水稻对干旱胁迫的敏感性提高。
7.2 bHLH转录因子bHLH家族是植物中仅次于MYB转录因子家族的第二大家族[103]。bHLH转录因子含有保守的bHLH结构域,该结构域高度保守,大约包含60个氨基酸,包含N端的碱性区和C端的HLH区。bHLH因子可以与顺式作用元件E-box(5'-CANNTG-3')特异性结合,其中又以G-box(5'-CACGTG-3')形式最为常见[104]。水稻中含有167个bHLH基因[105]。与植物其他转录因子一样,bHLH蛋白可能从渗透调节物质的积累、ROS平衡及激素信号等方面调控水稻的抗旱性。稻瘟病菌和渗透胁迫能诱导OsbHLH057表达。在PEG处理下,相比于野生型,OsbHLH057过表达植株中的SOD、POD和CAT活性显著升高而H2O2含量降低。进一步研究表明,OsbHLH057能与防卫相关基因Os2H16启动子区的AATCA顺式元件结合,从而提高调控水稻对稻瘟病、纹枯病及干旱胁迫的耐受性[106]。干旱也可以诱导OsbHLH130的表达,使得OsWIN2的表达量上升,从而促进蜡质合成基因的表达、降低失水速率和ROS的积累来增强水稻的抗旱性[107]。OsbHLH148的表达受茉莉酸(Jasmonic acid,JA)、ABA、干旱和低温等诱导。过量表达的OsbHLH148可诱导OsDREB和OsJAZ等基因的表达,并可通过OsbHLH148-OsJAZ1-OsCOI1途径共同影响JA信号通路,从而调控水稻的抗旱性[108]。
7.3 HD-ZIP蛋白HD-ZIP蛋白是植物特有的一类转录因子。它是由高度保守的同源异型结构域(Homeo domain,HD)和LZ组成,广泛参与植物胚胎发育、维管组织形成等生长发育过程及逆境胁迫应答。根据HD和LZ结构域的序列保守性和基因结构等特点,HD-ZIP蛋白分为4个亚家族:HD-ZIP Ⅰ、HD-ZIP Ⅱ、HD-ZIP Ⅲ和HD-ZIP Ⅳ[109]。目前,对HD-ZIP家族相关研究主要集中在双子叶模式植物拟南芥中,而在禾本科植物中的研究较少。HD-ZIP Ⅰ家族中的OsHOX22是负调控水稻抗旱性的因子,该基因可以结合AH2(CAAT(G/C)ATTG)顺式作用元件。研究表明,OsHOX22可以影响ABA的合成,通过ABA介导的信号传导途径参与调控干旱胁迫响应。过表达OsHOX22的转基因水稻植株体内ABA含量增加,对ABA的敏感性增强、对干旱胁迫的耐受性降低;相反,oshox22突变体中的ABA含量降低,在苗期抗旱性和耐盐性均得到增强[110]。类似地,OsHOX24也负向调控水稻的抗旱性。研究发现,在幼苗阶段,与野生型相比,过表达OsHOX24的水稻幼苗在干旱条件下苗长、根长均较短。在穗期,过高表达OsHOX24的水稻植株对干旱胁迫也更加敏感。通过转录组测序发现,其转录调控活性、碳水化合物、核酸和脂质代谢、响应非生物胁迫和激素信号等过程的相关基因发生明显变化,这暗示OsHOX24转录因子参与对下游代谢相关基因的调控[111]。与OsHOX22和OsHOX24不同,水稻HD-ZIP Ⅳ亚家族OsTF1L正向调控水稻的抗旱性。干旱条件下,高表达OsTF1L可以促进水稻光合作用,减缓其失水速率,从而显著提高水稻在营养生长阶段的抗旱性。在田间干旱条件下,与对照组相比,过表达OsTF1L植株在生殖生长阶段表现出更高的耐旱性。OsTF1L可以上调干旱诱导基因、气孔运动和木质素合成基因的表达量。此外,OsTF1L可以直接与木质素合成和干旱响应相关基因的启动子结合,比如poxN/PRX38(Class Ⅲ peroxidase)、DHHC4(DHHC-type domain containing protein)、CASPL5B1(Casparian Strip Membrane Domain Protein-Like Protein 5b1)等,从而共同参与干旱胁迫响应过程[112]。
7.4 MADS转录因子MADS的名称是由Minichromos memaintenance1(MCM1)、Agamous(AG)、Deficiens(DEF)及Serumresponse factor(SRF)这4个转录因子的首字母组成。MADS-box蛋白能特异性识别靶基因上游DNA核心序列CC[A/T]6GG,即CArG-box。MADS-box转录因子作为重要的调控因子参与调控植物生长和发育等过程。水稻基因组中包含约71个MADS转录因子[113],其中,OsMADS23受逆境诱导表达,其与SnRK2-型蛋白激酶SAPK9互作并将其磷酸化,进而增强OsMADS23的蛋白稳定性和转录活性。OsMADS23还可激活ABA和脯氨酸合成相关基因(OsNCED2、OsNCED3、OsNCED4和OsP5CR)的表达,影响内源ABA和脯氨酸的含量,从而正向调控水稻抗旱性[114]。与OsMADS23不同,OsMADS26负向调控水稻对稻瘟病、白叶枯病和干旱胁迫的抗性,Khong等[115]发现下调了水稻OsMADS26基因的表达量后,植株叶片的相对含水量明显提高,叶绿素含量下降速度减缓,且与胁迫相关的基因明显被诱导表达。同时,大田实验发现在干旱条件下,下调OsMADS26基因表达量的植株其产量明显高于野生型。
7.5 热应激蛋白和GRAS转录因子除以上转录因子外,目前也发现热应激蛋白和GRAS转录因子参与水稻干旱胁迫的调控过程。比如,过量表达热应激转录因子OsHsfA7能增强水稻苗期的抗旱性,相比于野生型,过量表达植株体内含有较低的叶片电导率和丙二醛含量[116]。GRAS转录因子家族这一命名来源于最初发现的3个家族成员,分别为Gibberellic acid insensitive(GAI)、Repressor of GA1-3 mutant(RGA)和Scarecrow(SCR)。研究表明,干旱、NaCl、JA均能诱导OsGRAS23的表达,高表达OsGRAS23能激活抗氧化相关基因的表达,从而提高转基因水稻对干旱和氧化应激的耐受性[117]。
8 结语与展望抗旱性是一种复杂的、多基因控制的数量性状。转录因子在干旱胁迫响应中发挥着重要作用。随着分子生物学和基因组学技术的发展,对转录因子的研究越发深入,为水稻抗旱育种提供了新的策略和方法。今后,可以在以下方面加强对转录因子在干旱胁迫响应中的研究:
(1)单个转录因子基因的过量表达或者敲除可能影响大量下游基因,而且可能会对不同的逆境胁迫及生长发育过程作出不同反应。比如,过量表达DREB2型转录因子OsDRAP1可以赋予转基因植株更高抗旱能力的同时,也会伴随着结实率降低和产量下降[65]。因此,在今后的研究中,可使用组织特异性启动子(如根特异性启动子)和受胁迫诱导启动子来实现不同转录因子基因在水稻中的应用。同时,可以加强构建水稻抗旱应答的转录调控网络,揭示不同转录因子之间的相互作用和协同效应,加强理解复杂的抗旱调控机制,最终促进它们在生产实际中的利用。
(2)2024年,《中共中央国务院关于学习运用“千村示范、万村整治”工程经验有力有效推进乡村全面振兴的意见》中提出要推动生物育种产业化扩面提速。由于通过基因编辑育种获得的品种完全不含外源基因,利用基因编辑技术进行作物改良育种的优势显而易见。例如,OsbZIP5转录因子被发现是水稻干旱胁迫耐受的负调节因子[11],可以通过CRISPR/Cas9等基因编辑技术敲除OsbZIP5转录因子,从而提高水稻的抗旱性,为未来作物改良提供可能性。因此,要加快基因编辑技术促进转录因子在水稻抗旱育种中的应用研究。
(3)随着生物信息学的不断发展,可以利用生物信息学工具和大数据分析,从全基因组水平上预测和验证新的转录因子,挖掘既能增加或者不影响产量等生长发育表型且能提高植物抗逆性的重要转录因子,研究它们在抗旱性调控中的潜在作用,为培育抗旱品种提供理论基础。
综上所述,转录因子在水稻抗旱性研究中具有巨大的潜力和广阔的前景。通过深入研究转录因子的功能和调控网络,结合现代生物技术,有望培育出更加高产、高抗的水稻新品种,为保障粮食安全和农业可持续发展做出贡献。
| [1] |
RAY D K, MUELLER N D, WEST P C, FOLEY J A, HART J P. Yield trends are insufficient to double global crop production by 2050[J]. PloS One, 2013, 8(6): e66428. DOI:10.1371/journal.pone.0066428 |
| [2] |
李洁. 植物干旱胁迫适应机制研究进展[J]. 广东农业科学, 2014, 41(19): 154-159. DOI:10.16768/j.issn.1004-874X.2014.19.005 LI J. Progress on drought stress adaptation mechanisms of plant[J]. Guangdong Agricultural Sciences, 2014, 41(19): 154-159. DOI:10.16768/j.issn.1004-874X.2014.19.005 |
| [3] |
WANG W S, QIN W Q, SUN F, WANG Y X, XU D D, LI Z K, FU B Y. Genome-wrde differences in DNA methylation changes in two ontrasting rice genotypes in response to drought conditionst[J]. Frontiers in Plant Science, 2016, 7: 1675. DOI:10.3389/fpls.2016.01675 |
| [4] |
KIM Y, CHUNG Y S, LEE E, TRIPATHI P, HEO S, KIM K H. Root response to drought stress in rice (Oryza sativa L.)[J]. International Journal of Molecular Sciences, 2020, 21(4): 1513. DOI:10.3390/ijms21041513 |
| [5] |
YANG S, VANDERBELD B, WAN J, HUANG Y. Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops[J]. Molecular Plant, 2010, 3(3): 469-490. DOI:10.1093/mp/ssq016 |
| [6] |
王彬, 陈敏氡, 林亮, 叶新如, 朱海生, 温庆放. 植物干旱胁迫的信号通路及相关转录因子研究进展[J]. 西北植物学报, 2020, 40(10): 1792-1806. DOI:10.7606/j.issn.1000-4025.2020.10.1792 WANG B, CHEN M D, LIN L, YE X R, ZHU H S, WEN Q F. Signal pathways and related transcription factors of drought stress in plants[J]. Acta Botanica Boreali-Occidentalia Sinica, 2020, 40(10): 1792-1806. DOI:10.7606/j.issn.1000-4025.2020.10.1792 |
| [7] |
李依雯, 张先文. 参与非生物胁迫的水稻bZIP转录因子家族分析[J/OL]. 分子植物育种, 1-13[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20230629.1450.004.html. LI Y W, ZHANG X W. Analysis of the rice bZIP transcription factor family involved in abiotic stress[J/OL]. Molecular Plant Breeding, 1-13[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20230629.1450.004.html. |
| [8] |
刘慧洁, 徐恒, 邱文怡, 李晓芳, 张华, 朱英, 李春寿, 王良超. bZIP转录因子在植物生长发育及非生物逆境响应的作用[J]. 浙江农业学报, 2019, 31(7): 1205-1214. DOI:10.3969/j.issn.1004-1524.2019.07.22 LIU H J, XU H, QIU W Y, LI X F, ZHANG H, ZHU Y, LI C S, WANG L C. Roles of bZIP transcription factors in plant growth and development and abiotic stress response[J]. Acta Agriculturae Zhejiangensis, 2019, 31(7): 1205-1214. DOI:10.3969/j.issn.1004-1524.2019.07.22 |
| [9] |
NIJHAWAN A, JAIN M, TYAGI A K, KHURANA J P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice[J]. Plant Physiology, 2008, 146(2): 333. DOI:10.1104/pp.107.112821 |
| [10] |
DAS P, LAKRA N, NUTAN KK, SINGLA-PAREEK SL, PAREEK A. A unique bZIP transcription factor imparting multiple stress tolerance in rice[J]. Rice, 2019, 12(1): 58. DOI:10.1186/s12284-019-0316-8 |
| [11] |
仝宇, 王聪, 赵利利, 连娟, 刘晓梅, 赵宝存. 转录因子OsbZIP5负调控水稻的耐旱性[J]. 中国生物化学与分子生物学报, 2021, 37(6): 798-810. DOI:10.13865/j.cnki.cjbmb.2021.03.1427 TONG Y, WANG C, ZHAO L L, LIAN J, LIU X M, ZHAO B C. Transcription factor OsbZIP5 negatively regulates drought-tolerance in rice[J]. Chinese Journal of Biochemistry and Molecular Biology, 2021, 37(6): 798-810. DOI:10.13865/j.cnki.cjbmb.2021.03.1427 |
| [12] |
ZOU M, GUAN Y, REN H, ZHANG F, CHEN F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance[J]. Plant Molecular Biology, 2008, 66(6): 675-683. DOI:10.1007/s11103-008-9298-4 |
| [13] |
JOO J, LEE Y H, SONG S I. Overexpression of the rice basic leucine zipper transcription factor OsbZIP12 confers drought tolerance to rice and makes seedlings hypersensitive to ABA[J]. Plant Biotechnology Reports, 2014, 8: 431-441. DOI:10.1007/s11816-014-0335-2 |
| [14] |
CHEN H, CHEN W, ZHOU J, HE H, CHEN L, CHEN H, DENG X W. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice[J]. Plant Science, 2012, 193/194: 8-17. DOI:10.1016/j.plantsci.2012.05.003 |
| [15] |
WANG B X, XU B, LIU Y, LI J F, SUN Z G, CHI M, XING Y G, YANG B, LI J, LIU J B, CHEN T M, FANG Z W, LU B G, XU D Y, BABATUNDE K B. A novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice (Oryza sativa L.)[J]. Plant Science, 2022, 323: 111393. DOI:10.1016/j.plantsci.2022.111393 |
| [16] |
XIANG Y, TANG N, DU H, YE H, XIONG L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice[J]. Plant Physiology, 2008, 148(4): 1938-1952. DOI:10.1104/pp.108.128199 |
| [17] |
ZONG W, TANG N, YANG J, PENG L, MA S, X U Y, LI G, XIONG L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]. Plant Physiology, 2016, 171(4): 2810-2825. DOI:10.1104/pp.16.00469 |
| [18] |
MA Y, WU Z, DONG J, ZHANG S, ZHAO J, YANG T, YANG W, ZHOU L, WANG J, CHEN J, LIU Q, LIU B. The 14-3-3 protein OsGF14f interacts with OsbZIP23 and enhances its activity to confer osmotic stress tolerance in rice[J]. The Plant Cell, 2023, 35(11): 4173-4189. DOI:10.1093/plcell/koad211 |
| [19] |
CHEN H, DAI X J, GU Z Y. OsbZIP33 is an ABA‐dependent enhancer of drought tolerance in rice[J]. Crop Science, 2015, 55(4): 1673-1685. DOI:10.2135/cropsci2014.10.0697 |
| [20] |
WU T, ZHANG M X, ZHANG H J, HUANG K, CHEN M J, CHEN C, YANG X, LI Z, CHEN H Y, MA Z M, ZHANG X M, JIANG W Z, DU X L. Identification and characterization of EDT1 conferring drought tolerance in rice[J]. Journal of Plant Biology, 2019, 62: 39-47. DOI:10.1007/s12374-018-0203-7 |
| [21] |
PARK S H, JEONG J S, LEE K H, KIM Y S, CHO Y D, KIM J K. OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance[J]. Plant Biotechnology Reports, 2015, 9: 89-96. DOI:10.1007/s11816-015-0346-7 |
| [22] |
TANG N, ZHANG H, LI X, XIAO J, XIONG L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice[J]. Plant Physiology, 2012, 158(4): 1755-1768. DOI:10.1104/pp.111.190389 |
| [23] |
LIU C, WU Y, WANG X. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice[J]. Planta, 2012, 235: 1157-1169. DOI:10.1007/s00425-011-1564-z |
| [24] |
喻旭, 牛向丽, 杨盛慧, 李欲翔, 刘亮亮, 唐维, 刘永胜. 过量表达转录因子OsbZIP60对水稻抗热和抗旱能力的研究[J]. 中国农业科学, 2011, 44(20): 4142-4149. DOI:10.3864/j.issn.0578-1752.2011.20.002 YU X, NIU X L, YANG S H, LI Y X, LIU L L, TANG W, LIU Y S. Research on heat and drought tolerance in rice (Oryza sativa L.) by overexpressing transcription factor OsbZIP60[J]. Scientia Agricultura Sinica, 2011, 44(20): 4142-4149. DOI:10.3864/j.issn.0578-1752.2011.20.002 |
| [25] |
YANG S, XU K, CHEN S, LI T, XIA H, CHEN L, LIU H, LUO L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice[J]. BMC Plant Biologyl, 2019, 19(1): 1-15. DOI:10.1186/s12870-019-1872-1 |
| [26] |
CHEN Y, SHEN J, ZHANG L, QI H, YANG L, WANG H, WANG J, WANG Y, DU H, TAO Z, ZHAO T, DENG P, SHU Q, QIAN Q, YU H, SONG S. Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice[J]. Molecular Plant, 2021, 14(8): 1297-1311. DOI:10.1016/j.molp.2021.05.001 |
| [27] |
LIU C, MAO B, OU S, WANG W, LIU L, WU Y, CHU C, WANG X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice[J]. Plant Molecular Biology, 2014, 84: 19-36. DOI:10.1007/s11103-013-0115-3 |
| [28] |
WANG B X, LIU Y, WANG Y F, LI J F, SUN Z G, CHI M, XING Y G, XU B, YANG B, LI J, LIU J B, CHEN T M, FANG Z W, LU B G, XU D Y, BABATUNDE K B. OsbZIP72 is involved in transcriptional gene-regulation pathway of abscisic acid signal transduction by activating rice high-affinity potassium transporter OsHKT1;1[J]. Rice Science, 2021, 28(3): 257-267. DOI:10.1016/j.rsci.2021.04.005 |
| [29] |
GAO W, LI M, YANG S, GAO C, SU Y, ZENG X, JIAO Z, XU W, ZHANG M, XIA K. miR2105 and the kinase OsSAPK10 co-regulate OsbZIP86 to mediate drought-induced ABA biosynthesis in rice[J]. Plant Physiology, 2022, 189(2): 889-905. DOI:10.1093/plphys/kiac071 |
| [30] |
金锋, 丁莲鑫, 骆骏, 聂圣松, 方中明. 水稻MYB转录因子的研究进展[J]. 植物遗传资源学报, 2023, 24(4): 917-926. DOI:10.13430/j.cnki.jpgr.20221220001 JIN F, DING L X, LUO J, NIE S S, FANG Z M. Research progress of MYB transcription factors in rice[J]. Journal of Plant Genetic Resources, 2023, 24(4): 917-926. DOI:10.13430/j.cnki.jpgr.20221220001 |
| [31] |
AMBAWAT S, SHARMA P, YADAV N R, YADAV R C. MYB transcription factor genes as regulators for plant responses: An overview[J]. Physiology and Molecular Biology of Plants, 2013, 19: 307-321. DOI:10.1007/s12298-013-0179-1 |
| [32] |
MUTHURAMALINGAM P, JEYASRI R, SELVARAJ A, SHIN H, CHEN J T, SATISH L, WU Q S, RAMESH M. Global integrated genomic and transcriptomic analyses of MYB transcription factor superfamily in C3 model plant Oryza sativa (L.) unravel potential candidates involved in abiotic stress signaling[J]. Frontiers in Ggenetics, 2022, 13: 946834. DOI:10.3389/fgene.2022.946834 |
| [33] |
YANG A, DAI X, ZHANG W H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice[J]. Journal of Experimental Botany, 2012, 63(7): 2541-2556. DOI:10.1093/jxb/err431 |
| [34] |
TANG Y, BAO X, ZHI Y, WU Q, GUO Y, YIN X, ZENG L, LI J, ZHANG J, HE W, LIU W, WANG Q, JIA C, LI Z, LIU K. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice[J]. Frontiers in Plant Science, 2019, 10: 168. DOI:10.3389/fpls.2019.00168 |
| [35] |
XIONG H, LI J, LIU P, DUAN J, ZHAO Y, GUO X, LI Y, ZHANG H, ALI J, LI Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice[J]. PLoS One, 2014, 9(3): e92913. DOI:10.1371/journal.pone.0092913 |
| [36] |
JIAN L, KANG K, CHOI Y, SUH M C, PAEK N C. Mutation of OsMYB60 reduces rice resilience to drought stress by attenuating cuticular wax biosynthesis[J]. The Plant Journal, 2022, 112(2): 339-351. DOI:10.1111/tpj.15947 |
| [37] |
QU X, ZOU J, WANG J, YANG K, WANG X, LE J. A rice R2R3-type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC[J]. International Journal of Molecular Sciences, 2022, 23(11): 5873. DOI:10.3390/ijms23115873 |
| [38] |
PENG Y, TANG N, ZOU J, RAN J, CHEN X B. Rice MYB transcription factor OsMYB1R1 negatively regulates drought resistance[J]. Plant Growth Regulation, 2023, 99(3): 515-525. DOI:10.1007/s10725-022-00922-w |
| [39] |
DAI X, XU Y, MA Q, XU W, WANG T, XUE Y, CHONG K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis[J]. Plant Physiology, 2007, 143(4): 1739-1751. DOI:10.1104/pp.106.094532 |
| [40] |
杨晨旭, 哈斯巴根, 杜超. 植物NAC转录因子结构和功能研究进展[J/OL]. 分子植物育种, 1-16[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20230403.1809.023.html. YANG C X, KHASBAGAN, DU C. Research progress on the structure and function of plant NAC transcription factors[J/OL]. Molecular Plant Breeding, 1-16[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20230403.1809.023.html. |
| [41] |
马雪祺, 阴艳红, 冯婧娴, 陈万生, 孙连娜, 肖莹. 植物NAC转录因子研究进展[J]. 植物生理学报, 2021, 57(12): 2225-2234. DOI:10.13592/j.cnki.ppj.2021.0338 MA X Q, YIN Y H, FENG J X, CHEN W S, SUN L N, XIAO Y. Research progress of NAC transcription factors in plant[J]. Plant Physiology Journal, 2021, 57(12): 2225-2234. DOI:10.13592/j.cnki.ppj.2021.0338 |
| [42] |
OOKA H, SATOH K, DOI K, NAGATA T, OTOMO Y, MURAKAMI K, MATSUBARA K, OSATO N, KAWAI J, CARNINCI P, HAYASHIZAKI Y, SUZUKI K, KOJIMA K, TAKAHARA Y, YAMAMOTO K, KIKUCHI S. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana[J]. DNA Research, 2003, 10(6): 239-247. DOI:10.1093/dnares/10.6.239 |
| [43] |
GENG A, LIAN W, WANG Y, LIU M, ZHANG Y, WANG X, CHEN G. Molecular mechanisms and regulatory pathways underlying drought stress response in rice[J]. International Journal of Molecular Sciences, 2024, 25(2): 1185. DOI:10.3390/ijms25021185 |
| [44] |
HU H, DAI M, YAO J, XIAO B, LI X, ZHANG Q, XIONG L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(35): 12987-12992. DOI:10.1073/pnas.0604882103 |
| [45] |
YOU J, ZONG W, LI X, NING J, HU H, LI X, XIAO J, XIONG L. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice[J]. Journal of Experimental Botany, 2013, 64(2): 569-583. DOI:10.1093/jxb/ers349 |
| [46] |
YOU J, ZONG W, HU H, LI X, XIAO J, XIONG L. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice[J]. Plant Physiology, 2014, 166(4): 2100-2114. DOI:10.1104/pp.114.251116 |
| [47] |
CHEN F, ZHANG H, LI H, LIAN L, WEI Y, LIN Y, WANG L, HE W, CAI Q, XIE H, ZHANG H, ZHANG J. IPA1 improves drought tolerance by activating SNAC1 in rice[J]. BMC Plant Biology, 2023, 23(1): 55. DOI:10.1186/s12870-023-04062-9 |
| [48] |
LEE D K, CHUNG P J, JEONG J S, JANG G, BANG S W, JUNG H, KIM Y S, HA S H, CHOI Y D, KIM J K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance[J]. Plant Biotechnology Journal, 2017, 15(6): 754-764. DOI:10.1111/pbi.12673 |
| [49] |
FANG Y, LIAO K, DU H, XU Y, SONG H, LI X, XIONG L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice[J]. Journal of Experimental Botany, 2015, 66(21): 6803-6817. DOI:10.1093/jxb/erv386 |
| [50] |
SHEN J, LV B, LUO L, HE J, MAO C, XI D, MING F. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice[J]. Scientific Reports, 2017, 7(1): 40641. DOI:10.1038/srep40641 |
| [51] |
JEONG J S, KIM Y S, REDILLAS M C, JANG G, JUNG H, BANG S W, CHOI Y D, HA S H, REUZEAU C, KIM J K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field[J]. Plant Biotechnology Journal, 2013, 11(1): 101-114. DOI:10.1111/pbi.12011 |
| [52] |
TAKASAKI H, MARUYAMA K, KIDOKORO S, ITO Y, FUJITA Y, SHINOZAKI K, YAMAGUCHI-SHINOZAKI K, NAKASHIMA K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice[J]. Molecular Genetics and Genomics, 2010, 284: 173-183. DOI:10.1007/s00438-010-0557-0 |
| [53] |
BANG S W, CHOI S, JIN X, JUNG S E, CHOI J W, SEO J S, KIM J K. Transcriptional activation of rice CINNAMOYL‐CoA REDUCTASE 10 by OsNAC5, contributes to drought tolerance by modulating lignin accumulation in roots[J]. Plant Biotechnology Journal, 2022, 20(4): 736-747. DOI:10.1111/pbi.13752 |
| [54] |
JEONG J S, KIM Y S, BAEK K H, JUNG H, HA S H, DO CHOI Y, KIM M, REUZEAU C, KIM J K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions[J]. Plant Physiology, 2010, 153(1): 185-197. DOI:10.1104/pp.110.154773 |
| [55] |
SHIM J S, OH N, CHUNG P J, KIM Y S, CHOI Y D, KIM J K. Overexpression of OsNAC14 improves drought tolerance in rice[J]. Frontiers in Plant Science, 2018, 9: 326760. DOI:10.3389/fpls.2018.00310 |
| [56] |
JUNG S E, KIM T H, SHIM J S, BANG S W, BIN YOON H, OH S H, KIM Y S, OH S J, SEO J S, KIM J K. Rice NAC17 transcription factor enhances drought tolerance by modulating lignin accumulation[J]. Plant Science, 2022, 323: 111404. DOI:10.1016/j.plantsci.2022.111404 |
| [57] |
HONG Y, ZHANG H, HUANG L, LI D, SONG F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice[J]. Frontiers in Plant Science, 2016, 7: 166009. DOI:10.3389/fpls.2016.00004 |
| [58] |
ZHENG X, CHEN B, LU G, HAN B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance[J]. Biochemical and Biophysical Research Communications, 2009, 379(4): 985-989. DOI:10.1016/j.bbrc.2008.12.163 |
| [59] |
CHEN X, WANG Y, LYU B, LI J, LUO L, LU S, ZHANG X, MA H, MING F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway[J]. Plant and Cell Physiology, 2014, 55(3): 604-619. DOI:10.1093/pcp/pct204 |
| [60] |
YUAN X, WANG H, CAI J, BI Y, LI D, SONG F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response[J]. BMC Plant Biology, 2019, 19(1): 1-19. DOI:10.1186/s12870-019-1883-y |
| [61] |
WANG B, WANG Y, YU W, WANG L, LAN Q, WANG Y, CHEN C, ZHANG Y. Knocking out the transcription factor OsNAC092 promoted rice drought tolerance[J]. Biology, 2022, 11(12): 1830. DOI:10.3390/biology11121830 |
| [62] |
HUANG L, HONG Y, ZHANG H, LI D, SONG F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance[J]. BMC Plant Biology, 2016, 16(1): 203. DOI:10.1186/s12870-016-0897-y |
| [63] |
黄前量, 徐庆国, 殷绪明. 水稻NAC转录因子基因OsDSR42的克隆与功能初步分析[J]. 分子植物育种, 2022, 20(16): 5229-5235. DOI:10.13271/j.mpb.020.005229 HUANG Q L, XU Q G, YIN X M. Cloning and functional primary analysis of OsDSR42, a NAC transcription factor gene from rice[J]. Molecular Plant Breeding, 2022, 20(16): 5229-5235. DOI:10.13271/j.mpb.020.005229 |
| [64] |
韩芳英, 胡昕, 王楠楠, 谢裕红, 王晓艳, 朱强. DREBs响应植物非生物逆境胁迫研究进展[J]. 生物技术通报, 2023, 39(11): 86-98. DOI:10.13560/j.cnki.biotech.bull.1985.2023-0124 HAN F Y, HU X, WANG N N, XIE Y H, WANG X Y, ZHU Q. Research progress in response of DREBs to abiotic stress in plant[J]. Biotechnology Bulletin, 2023, 39(11): 86-98. DOI:10.13560/j.cnki.biotech.bull.1985.2023-0124 |
| [65] |
HUANG L, WANG Y, WANG W, ZHAO X, QIN Q, SUN F, HU F, ZHAO Y, LI Z, FU B, LI Z. Characterization of transcription factor gene OsDRAP1 conferring drought tolerance in rice[J]. Frontiers in Plant Science, 2018, 9: 94. DOI:10.3389/fpls.2018.00094 |
| [66] |
CHAKRABORTY K, JENA P, MONDAL S, DASH G K, RAY S, BAIG M J, SWAIN P. Relative contribution of different members of OsDREB gene family towards osmotic stress tolerance in indica and japonica ecotypes of rice[J]. Plant Biology, 2021, 24(2): 356-366. DOI:10.1111/plb.13379 |
| [67] |
CHEN J Q, MENG X P, ZHANG Y, XIA M, WANG X P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice[J]. Biotechnology Letters, 2008, 30: 2191-2198. DOI:10.1007/s10529-008-9811-5 |
| [68] |
CUI M, ZHANG W, ZHANG Q, XU Z, ZHU Z, DUAN F, WU R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice[J]. Plant Physiolog y and Biochemistry, 2011, 49(12): 1384-1391. DOI:10.1016/j.plaphy.2011.09.012 |
| [69] |
MATSUKURA S, MIZOI J, YOSHIDA T, TODAKA D, ITO Y, MARUYAMA K, SHINOZAKI, YAMAGUCHI-SHINOZAKI K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes[J]. Molecular Genetics and Genomics, 2010, 283: 185-196. DOI:10.1007/s00438-009-0506-y |
| [70] |
KE Y G, YANG Z J, YU S W, LI TF, WU J H, GAO H, FU Y P, LUO L J. Characterization of OsDREB6 responsive to osmotic and cold stresses in rice[J]. Journal of Plant Biology, 2014, 57: 150-161. DOI:10.1007/s12374-013-0480-0 |
| [71] |
范永胜, 董彦琪, 张金霞, 朱红彩, 刘翼成, 屈涛, 朱坤, 杨海峰. WRKY转录因子在水稻非生物胁迫中的作用[J/OL]. 分子植物育种, 1-10[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20230820.1356.006.html. FAN Y S, DONG Y Q, ZHANG J X, ZHU H C, LIU Y C, QU T, ZHU K, YANG H F. The role of WRKY transcription factor in rice response to abiotic stress[J/OL]. Molecular Plant Breeding, 1-10[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20230820.1356.006.html. |
| [72] |
RAMAMOORTHY R, JIANG S Y, KUMAR N, VENKATESH P N, RAMACHANDRAN S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments[J]. Plant and Cell Physiology, 2008, 49(6): 865-879. DOI:10.1093/pcp/pcn061 |
| [73] |
SONG Y, JING S J, YU D Q. Overexpression of the stress-induced OsWRKY08 improves osmotic stress tolerance in Arabidopsis[J]. Chinese Science Bulletin, 2009, 54(24): 4671-4678. DOI:10.1007/s11434-009-0710-5 |
| [74] |
LEE H, CHA J, CHOI C, CHOI N, JI H S, PARK S R, LEE S, HWANG D J. Rice WRKY11 plays a role in pathogen defense and drought tolerance[J]. Rice, 2018, 11(1): 5. DOI:10.1186/s12284-018-0199-0 |
| [75] |
SHEN H, LIU C, ZHANG Y, MENG X, ZHOU X, CHU C, WANG X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice[J]. Plant Molecular Biology, 2012, 80(3): 241-253. DOI:10.1007/s11103-012-9941-y |
| [76] |
RAINERI J, WANG S, PELEG Z, BLUMWALD E, CHAN R L. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress[J]. Plant Molecular Biology, 2015, 88(4/5): 401-413. DOI:10.1007/s11103-015-0329-7 |
| [77] |
ZHANG M, ZHAO R, HUANG K, WEI Z, GUO B, HUANG S, LI Z, JIANG W, WU T, DU X. OsWRKY76 positively regulates drought stress via OsbHLH148-mediated jasmonate signaling in rice[J]. Frontiers in Plant Science, 2023, 14: 1168723. DOI:10.3389/fpls.2023.1168723 |
| [78] |
吕苗苗. 水稻逆境相关基因OsWRKY97和OsDUF946.4的功能分析[D]. 雅安: 四川农业大学, 2020. DOI: 10.27345/d.cnki.gsnyu.2020.000557. LYU M M. Functional analysis of stress related genes OsWRKY97 and OsDUF946.4 in rice[D]. Yaan: Sichuan Agricultural University, 2020. DOI: 10.27345/d.cnki.gsnyu.2020.000557. |
| [79] |
LIM C, KANG K, SHIM Y, YOO S C, PAEK N C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways[J]. Plant Physiology, 2022, 188(4): 1900-1916. DOI:10.1093/plphys/kiab492 |
| [80] |
TAO Z, KOU Y, LIU H, LI X, XIAO J, WANG S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice[J]. Journal of Experimental Botany, 2011, 62(14): 4863-4874. DOI:10.1093/jxb/err144 |
| [81] |
丁杰荣, 张静, 江立群, 吕树伟, 孙炳蕊, 李晨, 刘清. OsWRKY67负向调控水稻耐旱性的功能分析[J]. 分子植物育种, 2023, 21(10): 3272-3281. DOI:10.13271/j.mpb.021.003272 DING J R, ZHANG J, JIANG L Q, LYU S F, SUN B R, LI C, LIU Q. Functional analysis of OsWRKY67 in negatively regulating drought-tolerance in rice[J]. Molecular Plant Breeding, 2023, 21(10): 3272-3281. DOI:10.13271/j.mpb.021.003272 |
| [82] |
李琳, 丁峰, 潘介春, 张树伟, 黄幸, 王金英, 王颖, 李浩然, 徐炯志, 彭宏祥, 何新华. 植物锌指蛋白转录因子家族研究进展[J]. 热带农业科学, 2020, 40(2): 65-75. DOI:10.12008/j.issn.1009-2196.2020.02.011 LI L, DING F, PAN J C, ZHANG S W, HUANG X, WANG J Y, WANG Y, LI H R, XU J Z, PENG H X, HE X H. Research progress on family of plant zinc-finger protein transcription factors[J]. Chinese Journal of Tropical Agriculture, 2020, 40(2): 65-75. DOI:10.12008/j.issn.1009-2196.2020.02.011 |
| [83] |
XU D Q, HUANG J, GUO S Q, YANG X, BAO Y M, TANG H J, ZHANG H S. Overexpression of a TFⅢA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.)[J]. FEBS Letters, 2008, 582(7): 1037-1043. DOI:10.1016/j.febslet.2008.02.052 |
| [84] |
HUANG X Y, CHAO D Y, GAO J P, ZHU M Z, SHI M, LIN H X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control[J]. Genes & Development, 2009, 23(15): 1805-1817. DOI:10.1101/gad.1812409 |
| [85] |
LIU K, WANG L, XU Y, CHEN N, MA Q, LI F, CHONG K. Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice[J]. Planta, 2007, 226(4): 1007-1016. DOI:10.1007/s00425-007-0548-5 |
| [86] |
DANSANA P K, KOTHARI K S, VIJ S, TYAGI A K. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes[J]. Plant Cell Reports, 2014, 33(9): 1425-1440. DOI:10.1007/s00299-014-1626-3 |
| [87] |
SHARMA G, GIRI J, TYAGI A K. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis[J]. Plant Science, 2015, 237: 80-92. DOI:10.1016/j.plantsci.2015.05.011 |
| [88] |
KANNEGANTI V, GUPTA A K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice[J]. Plant Molecular Biology, 2008, 66: 445-462. DOI:10.1007/s11103-007-9284-2 |
| [89] |
WANG W, LIU B, XU M, JAMIL M, WANG G. ABA-induced CCCH tandem zinc finger protein OsC3H47 decreases ABA sensitivity and promotes drought tolerance in Oryza sativa[J]. Biochemical and Biophysical Research Communications, 2015, 464(1): 33-37. DOI:10.1016/j.bbrc.2015.05.087 |
| [90] |
ZHANG X, ZHANG B, LI M J, YIN X M. OsMSR15 encoding a rice C2H2-type zinc finger protein confers enhanced drought tolerance in transgenic Arabidopsis[J]. Journal of Plant Biology, 2016, 59: 271-281. DOI:10.1007/s12374-016-0539-9 |
| [91] |
LIU H, ZHANG H, YANG Y, LI G, YANG Y, WANG X, BASNAYAKE B M, LI D, SONG F. Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses[J]. Plant Molecular Biology, 2008, 68: 17-30. DOI:10.1007/s11103-008-9349-x |
| [92] |
ZENG D E, HOU P, XIAO F M, LIU Y S. Overexpressing a novel RING-H2 finger protein gene, OsRHP1, enhances drought and salt tolerance in rice (Oryza sativa L.)[J]. Journal of Plant Biology, 2014, 57: 357-365. DOI:10.1007/s12374-013-0481-z |
| [93] |
YUAN X, HUANG P, WANG R, LI H, LYU X, DUAN M, TANG H, ZHANG H, HUANG J. A zinc finger transcriptional repressor confers pleiotropic effects on rice growth and drought tolerance by down-regulating stress-responsive genes[J]. Plant and Cell Physiology, 2018, 59(10): 2129-2142. DOI:10.1093/pcp/pcy133 |
| [94] |
SAHID S, ROY C, SHEE D, SHEE R, DATTA R, PAUL S. ZFP37, C3H, NAC94, and bHLH148 transcription factors regulate cultivar-specific drought response by modulating r40C1 gene expression in rice[J]. Environmental and Experimental Botany, 2023, 214: 105480. DOI:10.1016/j.envexpbot.2023.105480 |
| [95] |
CUI L G, SHAN J X, SHI M, GAO J P, LIN H X. DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice[J]. PLoS Genetics, 2015, 11(10): e1005617. DOI:10.1371/journal.pgen.1005617 |
| [96] |
WU F, SHENG P, TAN J, CHEN X, LU G, MA W, HENG Y, LIN Q, ZHU S, WANG J, WANG J, GUO X, ZHANG X, LEI C, WAN J. Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice[J]. Journal of Experimental Botany, 2015, 66(1): 271-281. DOI:10.1093/jxb/eru417 |
| [97] |
KOTHARI K S, DANSANA P K, GIRI J, TYAGI A K. Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses[J]. Frontiers in Plant Science, 2016, 7: 198469. DOI:10.3389/fpls.2016.01057 |
| [98] |
葛奇, 席会鹏. 植物SPL转录因子研究进展[J]. 安徽农业科学, 2023, 51(23): 25-29, 53. DOI:10.3969/j.issn.0517-6611.2023.23.006 GE Q, XI H P. Research progress of SPL transcription factors in plants[J]. Journal of Anhui Agricultural Sciences, 2023, 51(23): 25-29, 53. DOI:10.3969/j.issn.0517-6611.2023.23.006 |
| [99] |
曾鑫海, 陈锐, 师宇, 盖超越, 范凯, 李兆伟. 植物SPL转录因子的生物功能研究进展[J]. 植物学报, 2023, 58(6): 982-997. DOI:10.11983/CBB22216 ZENG X H, CHEN R, SHI Y, GAI C Y, FAN K, LI Z W. Research advances in biological functions of plant SPL transcription factors[J]. Chinese Bulletin of Botany, 2023, 58(6): 982-997. DOI:10.11983/CBB22216 |
| [100] |
XIE K, WU C, XIONG L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice[J]. Plant Physiology, 2006, 142(1): 280-293. DOI:10.1104/pp.106.084475 |
| [101] |
曾慧玲. 转录因子OsSPL4在水稻干旱胁迫响应中的分子作用机理研究[D]. 福州: 福建农林大学, 2023. DOI: 10.27018/d.cnki.gfjnu.2023.000647. ZENG H L. Molecular mechanism of transcription factor OsSPL4 in response to drought stress in rice[D]. Fuzhou: FujianAgriculture and Forestry University, 2023. DOI: 10.27018/d.cnki.gfjnu.2023.000647. |
| [102] |
LI Y, HAN S, SUN X, KHAN N U, ZHONG Q, ZHANG Z, ZHANG H, MING F, LI Z, LI J. Variations in OsSPL10 confer drought tolerance by directly regulating OsNAC2 expression and ROS production in rice[J]. Journal of Integrative Plant Biology, 2023, 65(4): 918-933. DOI:10.1111/jipb.13414 |
| [103] |
蔡欣, 黄钰杏, 崔明月, 林明丽, 王一婷, 卢月, 王丽敏. 水稻OsbHLH109基因的表达分析[J]. 广东农业科学, 2023, 50(12): 120-130. DOI:10.16768/j.issn.1004-874X.2023.12.012 CAI X, HUANG Y X, CUI M Y, LIN M L, WANG Y T, LU Y, WANG L M. Expression analysis of OsbHLH109 gene in rice[J]. Guangdong Agricultural Sciences, 2023, 50(12): 120-130. DOI:10.16768/j.issn.1004-874X.2023.12.012 |
| [104] |
王昕嘉, 李昆志. 植物bHLH转录因子参与非生物胁迫信号通路研究进展[J]. 生命科学, 2015, 27(2): 208-216. DOI:10.13376/j.cbls/2015030 WANG X J, LI K Z. Progress of plant bHLH transcription factors involved in abiotic stress signaling pathways[J]. Chinese Bulletin of Life Sciences, 2015, 27(2): 208-216. DOI:10.13376/j.cbls/2015030 |
| [105] |
LI X, DUAN X, JIANG H, SUN Y, TANG Y, YUAN Z, GUO J, LIANG W, CHEN L, YIN J, MA H, WANG J, ZHANG D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiology, 2006, 141(4): 1167-1184. DOI:10.1104/pp.106.080580 |
| [106] |
LIU J, SHEN Y, CAO H, HE K, CHU Z, LI N. OsbHLH057 targets the AATCA cis-element to regulate disease resistance and drought tolerance in rice[J]. Plant Cell Reports, 2022, 41(5): 1285-1299. DOI:10.1007/s00299-022-02859-w |
| [107] |
GU X, GAO S, LI J, SONG P, ZHANG Q, GUO J, WANG X, HAN X, WANG X, ZHU Y, ZHU Z. The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice[J]. Plant Physiology and Biochemistry, 2021, 169: 269-279. DOI:10.1016/j.plaphy.2021.11.031 |
| [108] |
SEO J S, JOO J, KIM M J, KIM Y K, NAHM B H, SONG S I, CHEONG J J, LEE J S, KIM J K, CHOI Y D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. The Plant Journal, 2011, 65(6): 907-921. DOI:10.1111/j.1365-313X.2010.04477.x |
| [109] |
卢小云, 余礼, 张秀峰, 赵洁. 禾本科植物HD-ZIP转录因子研究进展[J]. 植物生理学报, 2021, 57(4): 727-738. DOI:10.13592/j.cnki.ppj.2021.0074 LU X Y, YU L, ZHANG X F, ZHAO J. Research progress of HD-ZIP transcription factor in gramineae plants[J]. Plant Physiology Journal, 2021, 57(4): 727-738. DOI:10.13592/j.cnki.ppj.2021.0074 |
| [110] |
ZHANG S, HAIDER I, KOHLEN W, JIANG L, BOUWMEESTER H, MEIJER A H, SCHLUEPMANN H, LIU C M, OUWERKERK P B. Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice[J]. Plant Molecular Biology, 2012, 80: 571-585. DOI:10.1007/s11103-012-9967-1 |
| [111] |
BHATTACHARJEE A, SHARMA R, JAIN M. Over-expression of OsHOX24 confers enhanced susceptibility to abiotic stresses in transgenic rice via modulating stress-responsive gene expression[J]. Frontiers in Plant Science, 2017, 8: 628. DOI:10.3389/fpls.2017.00628 |
| [112] |
BANG S W, LEE D K, JUNG H, CHUNG P J, KIM Y S, CHOI Y D, SUH J W, KIM J K. Overexpression of OsTF1L, a rice HD‐Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance[J]. Plant Biotechnology Journal, 2019, 17(1): 118-131. DOI:10.1111/pbi.12951 |
| [113] |
姚琦园, 李纷芬, 张林成, 周升恩. 植物MADS-box转录因子参与调控非生物胁迫的研究进展[J]. 江西农业学报, 2018, 30(5): 73-79. DOI:10.19386/j.cnki.jxnyxb.2018.05.15 YAO Q Y, LI F F, ZHANG L C, ZHOU S E. Research progress in MADS-box transcription factors involved in regulation of abiotic stress of plants[J]. Acta Agriculturae Jiangxi, 2018, 30(5): 73-79. DOI:10.19386/j.cnki.jxnyxb.2018.05.15 |
| [114] |
LI X, YU B, WU Q, MIN Q, ZENG R, XIE Z, HUANG J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice[J]. PLoS Genetics, 2021, 17(8): e1009699. DOI:10.1371/journal.pgen.1009699 |
| [115] |
KHONG G N, PATI P K, RICHAUD F, PARIZOT B, BIDZINSKI P, MAI C D, BÈS M, BOURRIÉ I, MEYNARD D, BEECKMAN T, SELVARAJ M G, MANABU I, GENGA A M, BRUGIDOU C, NANG DO V, GUIDERDONI E, MOREL J B, GANTET P. OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice[J]. Plant Physiology, 2015, 169(4): 2935-2949. DOI:10.1104/pp.15.01192 |
| [116] |
LIU A L, ZOU J, LIU C F, ZHOU X Y, ZHANG X W, LUO G Y, CHEN X B. Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice[J]. BMB Reports, 2013, 46(1): 31. DOI:10.5483/bmbrep.2013.46.1.090 |
| [117] |
XU K, CHEN S, LI T, MA X, LIANG X, DING X, LIU H, LUO L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes[J]. BMC Plant Biology, 2015, 15(1): 141. DOI:10.1186/s12870-015-0532-3 |
(责任编辑 马春敏)
 2024, Vol. 51
2024, Vol. 51