文章信息
基金项目
- 广东省乡村振兴战略专项资金种业振兴项目(2022-NYP-00-002,2022-NJS-00-004);国家重点研发计划(2021YFD1200101-05);广东省省长专项(粤财农〔2023〕145号);广东省农业科学院水稻研究所优谷计划(2022YG01);广东省水稻育种新技术重点实验室项目(2023B1212060042)
作者简介
- 邢俊连(1990—),女,在读博士生,研究方向为水稻抗旱种质资源鉴定评价与创新利用,E-mail:834430993@qq.com.
通讯作者
- 孙炳蕊(1980—),女,博士,副研究员,研究方向为作物遗传育种,E-mail:sunbingrui2003@163.com.
文章历史
- 收稿日期:2024-03-20
2. 华南农业大学农学院,广东 广州 510642
2. College of Agriculture, South China Agricultural University, Guangzhou 510642, China
我国干旱/半干旱区占国土面积的42%,在我国粮食生产中占有重要位置,全球气候变化和人类活动增加引发干旱事件频发[1]。水稻(Oryza sativa L.)作为世界主要粮食作物之一,为全球1/3以上人口提供主食,然而由于其根系小、角质层蜡薄和干旱胁迫下气孔迅速关闭而被认为是最易受干旱影响的作物之一[2-3]。据国家统计局数据显示,农业生产耗水量占我国淡水资源消耗总量的70%,而水稻种植耗水量占整个农业生产用水的70%,对水资源造成极大消耗,一旦发生旱灾将导致我国水稻严重减产。此外,水稻传统种植方式需长期保有水层,这会加重农药化肥的面源污染和温室气体排放,给环境带来严重挑战。面对淡水资源短缺现状,利用我国保存的丰富稻种资源进行抗旱性鉴定评价与创新利用,并培育出节水抗旱稻进行推广应用,符合国家可持续农业的发展战略。节水抗旱稻可旱播旱管、节水省工、便于机械化操作和改变传统种植方式,既能保证水稻优质高产,又可大量节约我国淡水资源。目前我国已培育出一批节水抗旱稻并进行了推广应用,为水稻抗旱育种和研究奠定良好基础。
水稻在长期进化过程中,形成一系列感知适应干旱逆境的有效机制来保证生命进程,包括激素含量调节、活性氧(Reactive oxygen species,ROS)清除、渗透调节、角质蜡沉积及根结构改善等。分子遗传学及组学等技术的进步,加速水稻抗旱研究,利用不同材料和群体通过关联作图、全基因组关联分析(Genome-wide association study,GWAS)、表达谱分析及组学分析等方法定位和挖掘出大量抗旱相关QTL位点和基因,并解析其参与抗旱的生理和分子机制,为水稻抗旱分子育种提供理论基础。
1 水稻抗旱性鉴定指标与品种选育植物抗旱性指在干旱环境条件下,仍可维持植株正常生长并保持产量的一种特性,根据抗旱机制分为逃旱性、避旱性、耐旱性和复水抗旱性4种类型[4]。逃旱性指通过调节发育进程避免干旱影响;避旱性指通过减少水分丢失和维持水分吸收保持细胞水势,如关闭气孔、减小叶面积、增大根系密度和深度等;耐旱性指通过自身遗传机制如渗透调节、增加细胞抗氧化能力及减少细胞体积等使其能够在低水势下维持一定膨压;复水抗旱性指遭受水分胁迫并复水后快速恢复生长的能力[4]。水稻抗旱性鉴定方法主要包括直接鉴定法和间接鉴定法两种,直接鉴定法是利用水稻种质在干旱胁迫条件下生长发育进程变化、生理变化及产量相关性状变化来鉴定水稻抗旱性的方法;间接鉴定法是利用生理和分子生物学等手段来鉴定水稻抗旱性[4]。水稻抗旱性鉴定最终体现在形态指标、生理生化指标及产量指标等。
1.1 形态指标1.1.1 叶形态指标 适当的卷叶和角质层阻力可减少叶面蒸腾,卷叶能减少叶面太阳辐射,起到保护自身的作用。研究表明,叶片大小、茸毛、蜡质、角质层厚度、气孔数量和开度等叶片形态特征可作为抗旱性鉴定指标[5],卷叶、枯叶和复水恢复能力等指标也能有效筛选苗期抗旱性品种[6]。国际水稻所在2013年修定的《水稻评价标准》中,将干旱胁迫后叶片卷曲程度、叶枯死率、复水恢复能力及结实率作为抗旱指标。
1.1.2 根系形态指标 健壮的根系具有良好的吸水能力,为植物在干旱条件下生长提供一定保障,旱稻的根一般具有长度较长、根基部较粗、根数少等特点[6-7]。一般情况下,粳稻通过广而深的根系系统应对干旱胁迫,而籼稻则通过缩短生长周期来规避干旱胁迫[8]。李其勇等[9]利用PEG-6000溶液模拟干旱对芽期种子萌发的影响,探讨芽期抗旱性鉴定指标并筛选抗旱材料,结果表明根长、根干重、根芽比等可用于鉴定芽期抗旱性。目前被广泛认可的根系形态指标是国际水稻所首创的根系拉力鉴定法,即水稻根系所需拉力越大根系就越发达,抗旱性也就越强[10]。因此,根生长角度、根数量、根长、根径、根干重、根密度、根系穿透力、维管束数目等均可作为抗旱鉴定指标。
1.2 生理生化指标在水分胁迫下,水稻植株的光合作用、呼吸作用、植物激素和生长调节因子等生理生化指标会发生变化以应对干旱环境、保护自身,因此可利用植物生理生化指标值鉴定水稻抗旱性,但目前尚未形成统一标准,多数情况下是以叶绿素含量、叶片水分相关指标、细胞质膜透性、酶活性、脯氨酸和脱落酸(Abscisic acid,ABA)含量及其他物质含量等作为评价水稻抗旱性的指标[11]。干旱胁迫条件下仍能保持较高叶绿素含量的水稻种质具有较强抗旱性;叶绿素a/b比值越大、游离脯氨酸含量越低、自由水含量低,品种的抗旱性越强[12]。
ABA调节细胞反应,使气孔关闭、蒸腾作用下降,从而减少水分蒸发。水分胁迫诱导ABA在植物根系中合成,并经导管向上运输到叶片后作用于气孔细胞使其关闭,ABA的积累增强植物体对干旱胁迫的抵抗能力[13]。脯氨酸吸湿性强,可增加植物体内束缚水含量来调节渗透压。干旱胁迫下,脯氨酸在植物体内大量积累,因此脯氨酸含量也在一定程度上反映了植物的抗旱能力[6]。
1.3 产量指标育种目的是培育品质好、产量高的品种,干旱胁迫条件下的高产稳产是节水抗旱稻育种的关键目标。与产量直接相关的农艺性状有穗粒数、有效穗数、千粒重及结实率等,因此这些农艺性状可作为产量指标用于抗旱性研究。在营养生长阶段,干旱胁迫会降低水稻有效穗数与生物量,从而导致减产;在抽穗开花期,干旱会导致结实率和千粒重降低[10]。Barnaby等[14]在探究干旱胁迫与产量间的关系中发现,干旱胁迫导致严重减产的品种气孔导度幅度增至最大、碳水化合物积累相对较低,表明在干旱胁迫下,水稻通过发生一系列生理生化变化影响产量。2016年农业农村部颁布的《节水抗旱稻抗旱性鉴定技术规范》通过测定产量及产量相关抗旱指数来判定抗旱性等级。
1.4 品种选育栽培稻有旱稻(Upland rice)和水稻(Lowland rice)两种生态型,旱稻通常在高海拔或干旱环境条件下生长,如云南、贵州等高海拔地区及海南干旱地区等,云南旱稻和海南山栏稻都是我国典型的旱稻资源。同时,我国从国际水稻所引进大量旱稻,并对其进行筛选和品种系统选育,育成‘中旱1号’‘中旱3号’。2005年,基于“水稻×旱稻”杂交育种的全球第一个优质旱稻不育系‘沪旱1A’问世,随后上海市农业生物基因中心陆续培育出‘旱优73’‘沪旱35’‘沪旱15’‘沪旱1516’等一系列优质节水抗旱稻并进行推广应用,‘旱优73’是长三角地区种植面积最大的杂交稻品种。节水抗旱稻既具有水稻的高产优质特性,又具有旱稻节水抗旱的特点,可实现旱种旱管的稻作生产模式,减少对水分的过度依赖及温室气体排放,促进水稻生产向资源节约型、环境友好型的可持续生产方式转型。2018年,经国家品种审定委员会批准,把节水抗旱稻作为一种特殊类型的水稻品种,启动国家区域试验,推动了节水抗旱稻的培育和推广应用。据国家水稻数据中心报道,截至目前已有40个节水抗旱稻获得新品种权,111个品种通过品种审定。
2 水稻抗旱生理机制 2.1 植物激素介导的信号调控机制植物激素在适应生物和非生物逆境中发挥着关键调节作用,参与水稻抗旱的植物激素主要有ABA、赤霉素(Gibberellin,GA)等。ABA不仅在种子休眠、种子萌发、根结构发育、胚成熟和气孔开闭过程中发挥重要作用,在应对干旱胁迫中也起到关键作用。ABA是气孔运动的主要调节因子,通过诱导叶片气孔快速关闭,防止蒸腾作用导致的水分流失[15]。目前已鉴定出许多参与ABA介导的应激信号通路蛋白,bZIP型转录因子通过与启动子中的G-BOX或ABA反应元件结合调控ABA响应基因的表达,从而触发一系列对环境刺激的反应[16],如OsGF14f通过与ABA应答转录因子OsbZIP23互作正向调节ABA应答,增强对下游靶基因的转录调节活性[17]。玉米GLK转录因子ZmGLK1和ZmG2作为叶绿体发育和生物发生的转录激活因子,在ABA介导下通过调节气孔快速闭合增强转基因水稻抗旱性[18]。转录因子OsERF71通过增强ABA信号传导和脯氨酸生物合成相关基因的表达增强水稻抗旱性[19]。过表达ABA胁迫-成熟诱导蛋白基因OsASR5可增加内源ABA水平和激活H2O2的生成,并诱导气孔关闭、降低气孔导度。此外,OsASR5作为伴侣样蛋白与热激蛋白HSP40和2OG-Fe(Ⅱ) 氧化酶互作,防止干旱胁迫相关蛋白失活[20]。ABA受体蛋白OsPYL/RCAR5影响ABA相关基因表达,进而调控气孔关闭以增强水稻抗旱性[21]。Pyrabactin Resistance(PYR)/Pyrabactin Resistance 1-like(PYL)/ABA受体的调控成分RCAR蛋白和2C型蛋白磷酸酶PP2Cs是ABA的受体和共受体[22],正常生长条件下,蛋白激酶SnRK2s被分支上的PP2Cs抑制;在干旱胁迫条件下ABA含量增加,当PYL感知到ABA时便形成PYL-ABA-PP2C复合物,然后释放、激活SnRK2s并使下游蛋白磷酸化[23]。水稻在干旱条件下体内GA含量降低,导致GA信号负调控因子SLENDER RICE 1积累,该因子通过竞争性结合后期促进复合物TAD1的共激活剂来抑制ABA受体PYL10降解,从而增强ABA反应和水稻的抗旱性[24]。
2.2 ROS清除机制ROS包括过氧化物阴离子、氢氧化物阴离子和H2O2,可调节植物生长发育、抵御生物和非生物胁迫。逆境条件下,植物体内ROS含量增多,在低浓度或中浓度下,ROS可作为应激信号通路中的第二信使触发应激防御反应,当ROS浓度积累到一定阈值后会导致脂类、DNA和蛋白质的氧化损伤甚至细胞死亡[25]。为平衡ROS含量,植物进化出包含抗氧化酶在内的抗氧化系统,抗氧化酶主要有超氧化物歧化酶(Superoxide dismutase,SOD)、过氧化氢酶(Catalase,CAT)、抗坏血酸过氧化物酶(Ascorbate peroxidase,APX)和非特异性过氧化物酶(Peroxidase,POD)等。
OsMRLK63是一种典型受体样激酶,作为快速碱化因子OsRALF45和OsRALF46的受体,可与几种NADPH氧化酶互作,从而磷酸化并调节ROS的产生以正向调节水稻抗旱性[26]。NADPH氧化酶基因OsRbohB的突变减少了细胞内ROS的产生,增强水稻对干旱的敏感性[27]。OsLG3通过调控下游抗氧化酶相关基因APX1、APX2、CATB、POD1、POD2和FeSOD的表达,促进ROS清除、提高水稻抗旱性[28]。OsMT1a过表达植株的CAT、POD和APX等活性显著增加,表明OsMT1a通过参与ROS清除通路增强水稻抗旱性[29]。OsESG1是一种受样体激酶,通过调节抗氧化酶活性和胁迫调节基因OsCAT和OsPOD的表达参与水稻抗旱响应[30]。OsSIK1是一种类受体激酶,通过激活抗氧化系统并影响叶表皮气孔密度在水稻抗旱过程中发挥作用[31]。OsSPL10、OsSPL14和OsSKIPa通过激活抗逆相关基因OsNAC2、SNAC1的表达,促进ROS清除和调控气孔闭合以增强水稻抗旱性[25, 32-33]。OsRLCK241通过增强ROS清除酶活性和渗透调节剂的积累来缓解干旱引起的渗透胁迫[34]。OsCBM1参与NADPH氧化酶介导的ROS产生,通过与OsRbohA和OsRacGEF1互作增强水稻抗旱性[35]。此外,通过对水稻转录组分析发现,抗旱品种的差异基因被显著富集在与抗氧化相关的代谢通路中[36]。由此可见,ROS清除机制一方面是干旱直接刺激抗氧化酶的产生,通过调节ROS含量参与抗旱;另一方面是通过蛋白与氧化酶互作,间接调节ROS含量参与抗旱。
2.3 渗透调节机制干旱条件下,水稻通过增加胞内渗透物质的浓度维持渗透压平衡,从而减缓或阻碍细胞脱水、保持正常细胞生理代谢,最终抵抗干旱胁迫。渗透物质分两类:从外部进入细胞的无机离子(K+、Na+、Mg2+、Cl-等)和内部合成的有机溶质(蔗糖、脯氨酸、酸菜碱、山梨醇等)[37]。
干旱抑制根系生长、影响K+吸收,导致K+积累减少,OsHAK1、OsAKT1和OsTPKb均为高亲和力的K+通道蛋白,OsHAK1通过调控RAA1表达促进水稻根系生长和K+积累,正向调节水稻抗旱性[38]。OsAKT1受CBL-CIP23复合体调控,可增强OsAKT1介导的K+吸收,过表达OsAKT1会增加根系中的K+水平,提高水稻对干旱胁迫的耐受性[39]。OsTPKb定位在小液泡的膜上,可改变小液泡K+状态,维持细胞内正常K+稳态,过量表达时可促进水稻在K+缺乏条件下正常生长,提高水稻对渗透和干旱胁迫的耐受性[40]。
脯氨酸是一种游离氨基酸,不仅是理想的渗透调节物质,而且可作为膜的保护物质及羟基自由基清除剂,在植物响应干旱胁迫进程中发挥作用[36]。研究表明,OsOLP1、OsTPS1、OsDHODH1、OsAMTR310、OsP5CS、OsERF65和OsCOIN均通过促进细胞内脯氨酸等渗透调节物质的积累来增强水稻抗旱性[37, 41-43]。
2.4 其他调控机制除以上调节机制外,水稻还可通过叶表面蜡质沉淀及根结构改善等机制抵御干旱胁迫。
叶表面蜡质是一种天然保护层,是植物抵御生物及非生物胁迫的重要屏障,能够减缓植物在干旱条件下非气孔水分的损失,维持植物体内水分平衡。OsGL1-1、OsGL1-2和OsGL1-3均为角质层蜡质合成相关蛋白,可调控水稻叶表面蜡质的合成和积累,进而提高角质层通透性、增强水稻对干旱的耐受性[44]。DHS是重要蜡质合成调控因子,具有E3泛素连接酶活性,转录因子ROC4可正向调控水稻表皮蜡质合成和干旱胁迫反应,DHS通过促进转录因子ROC4泛素化降解,从而负调控蜡质合成和水稻抗旱[45]。OsMYB60通过与蜡质生物合成关键调节因子OsCER1的启动子结合,促进叶表面角质蜡生物合成,增强水稻抗干旱胁迫能力[46]。DWA1在水稻维管组织和表皮层优先表达,受干旱胁迫诱导,dwa1突变体中与角质蜡合成相关基因的表达显著受抑制,说明DWA1通过调控角质蜡沉积控制水稻抗干旱能力[47]。
RRS1编码一种负调控根发育的R2R3型MYB转录因子,通过诱导OsIAA的表达抑制根发育;敲除RRS1可促进水稻根系发育,进而促进水分吸收、提高水分利用率,最终增强水稻抗旱性[48]。OsERF71通过上调木质素合成基因和细胞壁松弛相关基因的表达促进根部发育、增强水稻抗旱性[42]。DRO1可调控水稻根系生长角度,并受生长素负调控,参与根尖细胞伸长,过量表达该基因会增加根系生长角度使根系向更下方生长,通过控制根系结构增强水稻抗旱性[49]。WOX11可与ERF3互作,并通过促进根毛生长、侧根初始化和冠根伸长提高水稻抗旱性[50]。
3 水稻抗旱性QTL定位与基因挖掘 3.1 水稻抗旱性QTL定位水稻抗旱性是由多基因控制的数量性状,受环境因素影响较大,且表现为连续变异。低密度的遗传图谱和不稳定的表型性状,是进行遗传分析和挖掘抗旱基因的障碍。借助分子标记技术和高通量测序技术,可在分子水平表征数量庞大的种质资源,评估抗旱资源多样性,鉴定出抗旱性强、品质优的资源供育种利用。目前通过不同作图群体和抗旱表型定位到大量与水稻抗旱性相关QTL,这有助于挖掘抗旱基因、解析抗旱机制。最早报道定位到水稻抗旱性QTL的是Chmpoux等[51],其利用籼稻和粳稻杂交获得重组自交系群体,绘制了包含127个RFLPs标记的连锁图谱,得到45个与根粗、根冠比、根干重、最大根长等性状的相关QTL。此后,陆续有干旱相关的QTL通过分子标记技术被鉴定到[52]。Satrio等[53]利用IR64和Hawara Bunar衍生的重组自交系,构建了高密度的SNP标记,检测到分别位于1、3、6、8、9、12号染色体上13个区域的41个QTL,其中3个热点QTL位于6号和8号染色体上,2个主要QTL位于9号染色体上。水稻在生殖生长阶段遭遇干旱胁迫会导致严重减产,Bhattarai等[54]利用Cocodrie和N-22重组自交系群体绘制了包括4 747个SNP标记的遗传连锁图谱,在干旱胁迫下共检测到21个与开花、株高、卷叶程度、干物质含量、结实率、产量、产量系数及收获系数相关QTL,大效应QTL(qDTF3.01、qPH1.38、qLRS1.37)和产量QTL(qGYS1.42)可与优良育种系结合培育节水抗旱稻品种。Barik等[55]利用CR 143-2-2和Krishnahamsa重组自交系群体190 F7绘制了基于SSR标记的遗传连锁图谱,在水稻生殖生长阶段检测到3个新的QTL并分别调控相对叶绿素含量、叶绿素含量及脯氨酸含量,这3个QTL将有助于利用标记辅助育种方法提高水稻在生殖生长阶段的抗旱性。Sun等[56]利用自然群体进行连锁分析及GWAS定位到多个水稻抗旱相关QTL,克隆出多个具有重要育种价值的抗旱基因,如OsLG3、DROT1和RRS1等。
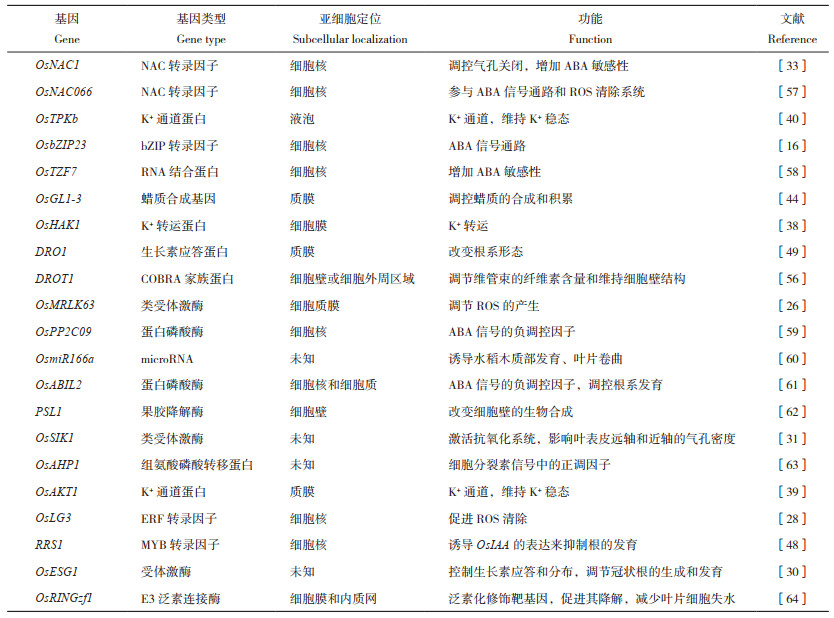
3.2 水稻抗旱基因目前,水稻中已有分布在12条染色体上的378个抗旱基因被克隆(国家水稻数据中心),这些抗旱基因多为转录因子、转运蛋白、泛素化酶、磷酸酶和蛋白激酶等,并从已报道的各类型基因中挑取1~2个列于表 1中。
3.2.1 转录因子 转录因子(Transcription factors,TFs)在植物逆境信号传递过程中起重要调节作用,能调控抗逆相关基因表达,从而增强植物抗逆性。干旱胁迫过程中,植物感受到干旱信号后会发生一系列生理生化的改变,进而通过一系列信号传递激活转录因子,被激活的转录因子可与靶基因的顺式作用元件特异性结合,启动下游特定应答基因转录表达,从而作出对干旱的应激反应[65]。目前发现水稻中参与干旱胁迫应答的转录因子主要有NAC转录因子、WRKY转录因子、MYB转录因子、bZIP转录因子、AP2/ERF转录因子、DREB转录因子和TIFY转录因子等[66]。
NAC转录因子超家族是最重要的植物特异性转录因子家族之一,拥有150多个成员,在N端NAC结构域高度保守,可与DNA结合;具有高度可变的C端结构域用于转录激活,在涉及器官发育、二次细胞壁合成、衰老和相应非生物胁迫的各种过程中都是至关重要的调节因子[67]。OsNAC5、OsNAC6、OsNAC10、OsNAC14、OsNAC066均通过增强参与根系结构和胁迫反应基因的表达,提高水稻抗旱性[57, 68-71],OsNAC17通过调节木质素积累增强水稻抗旱性[67],而OsNAC095负调节水稻抗旱性,正调节水稻耐冷性[72]。
植物特异性WRKY转录因子拥有WRKY DNA结合域[73],在水稻生长发育及抵御非生物胁迫和生物胁迫过程中具有重要调控作用,且大多数WRKY转录因子参与植物免疫反应[74]。水稻具有103个WRKY转录因子,其中OsWRKY5、OsWRKY15、OsWRKY45和OsWRKY114可负调控水稻抗旱性,OsWRKY11、OsWRKY30和OsWRKY47正调控水稻的抗旱性[75]。
AP2/ERF超基因家族编码的蛋白质由AP2/ERF结构域定义,该结构域由50~60个氨基酸组成,这些蛋白质参与植物生命周期中的各种调节机制,水稻中共鉴定出170多个AP2/ERF基因,但大部分基因功能有待确定[76-77]。ERF亚家族基因的表达在不同物种中由多种应激反应诱导,并在应激和激素应答信号通路中作为中枢蛋白发挥作用[78]。干旱胁迫下,OsERF48和OsERF71在根系中优先表达,通过促进根系发育赋予水稻抗旱性[79-81]。OsERF71通过上调ABA响应基因OsABI5、OsPP2C68、OsRAB16A和OsRAB16C及脯氨酸合成基因OsP5CS1和OsP5CS2的表达,促进ABA的合成和脯氨酸的积累以提高水稻抗旱能力[19]。敲除OsEBP89可改善浸水条件下的种子萌发,并增强水稻抗旱性[82]。在水稻生殖期进行干旱胁迫,当过量表达OsERF101时可提高植株结实率[83]。OsERF83定位于细胞核,过表达该基因可增强水稻抗旱性[65]。
MYB转录因子指含MYB结构域的转录因子,该结构域在真核生物中高度保守,作为反式作用结构域参与MYB蛋白功能的调节[84]。水稻MYB转录因子家族包含155个MYB基因[85],已有研究表明,MYB转录因子参与水稻抗旱调控:OsMYB6和OsMYB105定位在细胞核,过量表达后可提高水稻抗旱性[86-87];R2R3-MYB转录因子ScMYBAS1可正向调节水稻抗旱性[88];过表达OsMYBR1可降低ABA敏感性并提高水稻抗旱性[89];过表达OsMYB48-1可增强水稻抗旱性和耐盐性[90];SiMYB56通过调节木质素生物合成和ABA信号通路,从而增强水稻抗旱性[91];OsMYBR57直接调节关键干旱胁迫应答基因OsbZIPs表达,并与同源结构域转录因子OsHB22互作协同调控干旱相关基因的表达[92];R2R3-MYB转录因子OsFLP主要通过调节OsNAC1和OsNAC6的转录来应对干旱胁迫[93]。
bZIP家族转录因子在植物抗逆调控通路中发挥着重要调控作用,如OsbZIP5是水稻干旱胁迫耐受的负调节因子[94];OsbZIP62能结合到OsNAC10、DSM2等抗逆相关基因的上游启动子上,促进其上调表达,从而增强水稻对干旱胁迫的耐受性[95]。
3.2.2 转运蛋白 K+通道蛋白是一种跨膜转运蛋白,广泛存在于各种细胞膜上,通过控制通道的开关维持细胞内的离子稳态,在水稻抗旱过程中发挥作用。例如,前文提到的OsHAK1、OsAKT1、OsTPKb[38-40]。AM1编码K+外排逆向转运蛋白,可调控叶绿素生物合成,通过积累H2O2和关闭气孔增强水稻抗旱能力[96]。OsDIL编码一个脂质转运蛋白,干旱条件下OsDIL过表达植株的脯氨酸和可溶性糖含量升高,SOD和POD活性也明显升高,花粉育性和产量提高[97]。OsNHX1是Na+/H+逆向转运蛋白基因,定位在液泡膜上,可将胞质中高浓度的Na+和K+隔离,通过渗透调节提高水稻抗旱性[98]。
3.2.3 泛素化酶 泛素化酶指一类与泛素化修饰相关酶,包括泛素激活酶、泛素结合酶和泛素连接酶3种,水稻中已克隆的泛素化基因主要是泛素连接酶基因,这些基因参与蛋白质泛素化过程,通过泛素化介导靶基因降解提高水稻抗旱。OsRINGzf1定位于细胞膜和内质网上,可与水通道蛋白OsPIP2;1互作并进行泛素化修饰,促进其降解,以减少叶片细胞失水,进而调控水稻抗旱[64]。OsREIW1可泛素化OsWRKY31、促进其降解,功能缺失的突变体osrdcp1在正常和干旱胁迫下与野生型无明显差异,而过表达植株则表现出抗旱性[99]。OsRF1定位于内质网膜上,靶向OsPP2C09蛋白并对其进行泛素化降解,此外,过量表达OsRF1可上调ABA生物合成基因的表达、提高ABA水平、降低其失水率,从而增强水稻的抗旱性及耐盐性[100]。泛素化酶OsETOL1与OsACS2互作,通过调节乙烯含量和能量代谢在水稻干旱胁迫中发挥作用[101]。OsPUB16定位于细胞膜和细胞核,通过抑制ABA和茉莉酸(Jasmonic acid,JA)生物合成,调节“SAPK9-OsMADS23-OsAOC”途径降低水稻对缺水的耐受能力,OsMADS23与启动子结合激活JA生物合成基因OsAOC的表达,SAPK9介导的OsMADS23磷酸化通过干扰OsPUB16-OsMADS23相互作用降低泛素化水平,从而增强OsMADS23的稳定性并促进OsAOC的表达[102]。泛素化酶基因OsDIRP1定位于细胞核,负调控水稻抗旱能力,正调控水稻耐寒性[104]。泛素化酶基因OsCLR1通过提高ABA含量正向调控水稻耐盐性和干旱胁迫应答[104]。
3.2.4 蛋白激酶和磷酸酶 蛋白磷酸化和去磷酸化过程在多种信号识别和转导中起重要作用,水稻中已发现多种蛋白激酶和磷酸酶基因参与到水稻抗旱进程。DSM1编码一个有丝分裂原活化蛋白激酶,定位在细胞核,作为水稻响应干旱胁迫的早期信号传导组分,通过调节POD基因表达控制ROS清除,从而调节水稻抗旱性[105]。蛋白激酶SAPK2定位于细胞核和细胞质,是ABA信号通路的正调控因子,与OsbZIP23互作并将其磷酸化,从而激活OsbZIP23的转录激活活性。磷酸酶OsPP2C49与SAPK2互作,使SAPK2失活以抑制OsbZIP23的转录激活活性,从而负调控ABA信号通路,且OsbZIP23可与OsPP2C49启动子结合,正调控OsPP2C49表达[16]。OsPP18是一个受SNAC1调控的蛋白磷酸酶,定位在细胞质和细胞核,通过不依赖ABA的ROS清除途径调控水稻对干旱和氧化胁迫的抗性[106]。OsPP2C09是一种蛋白磷酸酶,定位于细胞核,OsRF1表现出E3连接酶活性,通过靶向OsPP2C09蛋白进行泛素化降解提高水稻抗旱能力[100]。OsPFA-DSP1是一种蛋白酪氨酸磷酸酶,定位在细胞质和细胞核,负调控转基因水稻的干旱胁迫反应[107]。
尽管水稻中越来越多的抗旱相关基因被发现和克隆,并证实可在水稻抗旱性中发挥作用,但它们参与干旱胁迫反应的机制及彼此之间的互作关系尚未完全阐明,且这些基因很少能为育种家所用,值得我们进一步去探索和利用。
4 结语与展望水稻生产用水占全国总耗水量的50%,为适应我国水资源短缺的严峻形势、确保国家粮食安全,我国在节水抗旱稻品种选育上取得重大进展。例如,上海市农业生物基因中心育成一批优质节水抗旱稻并推广应用,其中‘旱优73’在长三角地区得到广泛种植[108]。国家行业标准《节水抗旱稻抗旱性鉴定技术规范》和国家区域试验规范了抗旱性鉴定流程,推动了节水抗旱稻的培育和推广应用,加快了水稻生产方式向资源节约型、环境友好型的可持续生产方式转变。但通过审定的品种数量远不能满足水资源匮乏的水稻主产区对节水抗旱稻品种的迫切需要,适应于不同生态区域的品种更为匮乏。此外,这些品种选育侧重于抗旱性的提升,产量优势不强。
种质资源是国家战略性资源,事关种业振兴全局,我国收集保存了丰富的水稻种质资源,同时也引进大批旱稻资源,应加强对种质资源尤其是野生稻资源和农家品种的鉴定评价和创新利用[109-110]。利用形态指标和生理生化指标筛选鉴定出一些强抗旱性种质,但最终都要以产量和品质作为鉴定指标。目前,旱稻资源的筛选多集中于水稻苗期,应加强对水稻全生育期的抗旱鉴定,从根本上解决育种家的实际需求,筛选出可直接作为旱稻品种选育的优良亲本。
近年来,水稻抗旱性的生理和分子机制得到广泛研究,渗透调节机制、ROS清除机制、植物激素调节机制、蜡质沉积及根结构改善均在水稻抗旱中起到重要作用。利用不同材料和群体通过关联作图、GWAS、表达谱分析及多组学联合分析定位并克隆出很多与水稻抗旱相关QTL和基因,这些基因多与转录因子、转运蛋白、泛素化酶、蛋白激酶和磷酸酶等有关。除OsLG3、DROT1、RRS1等几个主效基因是利用正向遗传学方法克隆得到,其他相关基因均利用反向遗传学的方法克隆获得,并不一定是真正意义上的抗旱基因,基因间的互作关系和抗旱机制仍有待于进一步验证和探索。此外,这些抗旱相关基因鲜少被育种家利用,基础研究要与应用研究相结合,充分利用这些真正的抗旱基因来培育出更优质的节水抗旱稻品种。利用分子标记辅助育种和遗传修饰育种可定向、高效培育优良作物,大幅加快育种效率,缩短育种周期,但目前尚无通过分子育种手段育成水稻抗旱品种的报道。
| [1] |
符淙斌, 马柱国. 全球干旱/半干旱区年代尺度干湿变化研究的进展及思考[J]. 大气科学学报, 2023, 46(4): 481-490. DOI:10.13878/j.cnki.dqkxxb.20230517001 FU C B, MA Z G. Progress and reflection on the study of decadal dry and wet changes in global arid/semi arid regions[J]. Transactions of Atmospheric Sciences, 2023, 46(4): 481-490. DOI:10.13878/j.cnki.dqkxxb.20230517001 |
| [2] |
SAHEBI M, HANAFI M M, RAFⅡ M Y, MAHMUD T, AZIZI P, OSMAN M, ABIRI R, TAHERI S, KALHORI N, SHABANIMOFRAD M, MIAH G, ATABAKI N. Improvement of drought tolerance in rice (Oryza sativa L.) genetics, genomic tools, and the WRKY gene family[J]. Biomed Research International, 2018, 3158474. DOI:10.1155/2018/3158474 |
| [3] |
OLADOSU Y, RAFⅡ M Y, SAMUEL C, FATAI A, MAGAJI U, KAREEM I, KANARUDIN Z S, MUHAMMAD I, KOLAPO K. Drought resistance in rice from conventional to molecular breeding: A review[J]. International Journal of Molecular Sciences, 2019, 20(14): 3519. DOI:10.3390/ijms20143519 |
| [4] |
马孝松. 稻种资源抗旱性评价、基因发掘与耐旱机制研究[D]. 武汉: 华中农业大学, 2017. DOI: 10.7666/d.Y3217509. MA X S. Drought resistance evaluation, gene discovery and study on drought tolerance mechanism in rice[D]. Wuhan: Huazhong Agricultural University, 2017. DOI: 10.7666/d.Y3217509. |
| [5] |
胡标林, 李名迪, 万勇, 朱雪晶, 张铮. 我国水稻抗旱性鉴定方法与指标研究进展[J]. 江西农业学报, 2005(2): 56-60. DOI:10.19386/j.cnki.jxnyxb.2005.02.015 HU B L, LI M D, WAN Y, ZHU X J, ZHANG Z. Advances in identification methods and index of rice resistance to drought in China[J]. Acta Agriculturae Jiangxi, 2005(2): 56-60. DOI:10.19386/j.cnki.jxnyxb.2005.02.015 |
| [6] |
马廷臣, 余蓉蓉, 陈荣军, 曾汉来, 张端品. PEG-6000模拟干旱对水稻幼苗期根系的影响[J]. 中国生态农业学报, 2010, 18(6): 1206-1211. DOI:10.3724/SP.J.1011.2010.01206 MA T C, YU R R, CHEN R J, ZENG H L, ZHANG D P. Effect of drought simulated with PEG-6000 on shoot system in rice seeding[J]. Chinese Journal of Eco-Agriculture, 2010, 18(6): 1206-1211. DOI:10.3724/SP.J.1011.2010.01206 |
| [7] |
马一泓, 王术, 于佳禾, 赵晨, 贾宝艳, 黄元财, 王岩, 王韵, 徐铨. 水稻生长对干旱胁迫的响应及抗旱性研究进展[J]. 种子, 2016, 35(7): 45-49. DOI:10.16590/j.cnki.1001-4705.2016.07.045 MA Y H, WANG S, YU J H, ZHAO C, JIA B Y, HUANG Y C, WANG Y, WANG Y, XU Q. Response of drought stress in plant growth and study advances on drought resistance in rice[J]. Seed, 2016, 35(7): 45-49. DOI:10.16590/j.cnki.1001-4705.2016.07.045 |
| [8] |
KIM Y, CHUNG Y S, LEE E, TRIPATHI P, HEO S, KIM K H. Root response to drought stress in rice (Oryza sativa L.)[J]. International Journal of Molecular Sciences, 2020, 21(4): 1513. DOI:10.3390/ijms21041513 |
| [9] |
李其勇, 朱从桦, 李星月, 向运佳, 杨晓蓉, 符慧娟, 张鸿. 水稻近等基因导入系芽期抗旱性鉴定及抗旱指标筛选[J]. 核农学报, 2021, 35(1): 192-201. DOI:10.11869/j.issn.100-8551.2021.01.0192 LI Q Y, ZHU C H, LI X Y, XIANG Y J, YANG X R, FU H J, ZHANG H. Drought resistance identification and index screening of rice near-isogenic introgression lines at germinating stage[J]. Journal of Nuclear Agricultural Sciences, 2021, 35(1): 192-201. DOI:10.11869/j.issn.100-8551.2021.01.0192 |
| [10] |
饶玉春, 戴志俊, 朱怡彤, 姜嘉骥, 马若盈, 王予烨, 王跃星. 水稻抗干旱胁迫的研究进展[J]. 浙江师范大学学报(自然科学版), 2020, 43(4): 417-429. DOI:10.16218/j.issn.1001-5051.2020.04.009 RAO Y C, DAI Z J, ZHU Y T, JIANG J J, MA R Y, WANG Y Y, WANG Y X. Advances in research of drought resistance in rice[J]. Journal of Zhejiang Normal University (Natural Sciences), 2020, 43(4): 417-429. DOI:10.16218/j.issn.1001-5051.2020.04.009 |
| [11] |
DEIVANAI S. Physiochemical traits as potential indicators for determining drought tolerance during active tillering stage in rice (Oryza sativa L.)[J]. Pertanika Journal of Tropical Agricultural Science, 2010, 43(4): 417-429. DOI:10.1007/978-1-4302-5370-9 |
| [12] |
程建峰, 潘晓云, 刘宜柏, 戴廷波, 曹卫星. 快速鉴定稻种资源抗旱性的生理指标筛选及其遗传背景[J]. 西南农业学报, 2005(5): 529-533. DOI:10.16213/j.cnki.scjas.2005.05.006 CHENG J F, PAN X Y, LIU Y B, DAI T B, CAO W X. Physiological index of rapid identification for drought resistance of rice germplasms and their genetic backgrounds[J]. Southwest China Journal of Agricultural Sciences, 2005(5): 529-533. DOI:10.16213/j.cnki.scjas.2005.05.006 |
| [13] |
范晓荣, 沈其荣. ABA、IAA对旱作水稻叶片气孔的调节作用[J]. 中国农业科学, 2003(12): 1450-1455. DOI:10.3321/j.issn:0578-1752.2003.12.005 FAN X R, SHEN Q R. Effects of ABA and IAA on the behavior of stomata of rice crop cultivated in aerobic soil condition[J]. Scientia Agricultura Sinica, 2003(12): 1450-1455. DOI:10.3321/j.issn:0578-1752.2003.12.005 |
| [14] |
BARNABY J Y, ROHILA J S, HENRY C G, SICHER R C, REDDY V R, MCCLUNG A M. Physiological and metabolic responses of rice to reduced soil moisture: Relationship of water stress tolerance and grain production[J]. International Journal of Molecular Sciences, 2019, 20(8): 1846. DOI:10.3390/ijms20081846 |
| [15] |
HSU P K, DUBEAUX G, TAKAHASHI Y, SCHROEDER J I. Signaling mechanisms in abscisic acid-mediated stomatal closure[J]. The Plant Journal, 2021, 105(2): 307-321. DOI:10.1111/tpj.15067 |
| [16] |
ZONG W, TANG N, YANG J, PENG L, MA S, XU Y, LI G, XIONG L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]. Plant Physiology, 2016, 171(4): 2810-2825. DOI:10.1104/pp.16.00469 |
| [17] |
MA Y, WU Z, DONG J, ZHANG S, ZHAO J, YANG T, YANG W, ZHOU L, WANG J, CHEN J, LIU Q, LIU B. The 14-3-3 protein Osgf14F interacts with Osbzip23 and enhances its activity to confer osmotic stress tolerance in rice[J]. Plant Cell, 2023, 35(11): 4173-4189. DOI:10.1093/plcell/koad211 |
| [18] |
LI X, LI J, WEI S, GAO Y, PEI H, GENG R, LU R, WANG P, ZHOU W. Maize golden2-like proteins enhance drought tolerance in rice by promoting stomatal closure[J]. Plant Physiology, 2024, 194(2): 774-786. DOI:10.1093/plphys/kiad561 |
| [19] |
LI J, GUO X, ZHANG M, WANG X, ZHAO Y, YIN Z, ZHANG Z, WANG Y, XIONG H, ZHANG H, TODOROVSKA E, LI Z. OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis[J]. Plant Science, 2018, 270: 131-139. DOI:10.1016/j.plantsci.2018.01.017 |
| [20] |
LI J, LI Y, YIN Z, JIANG J, ZHANG M, GUO X, YE Z, ZHAO Y, XIONG H, ZHANG Z, SHAO Y, JIANG C, ZHANG H, AN G, PEAK N C, ALI J, LI Z. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice[J]. Plant Biotechnology Journal, 2017, 15(2): 183-196. DOI:10.1111/pbi.12601 |
| [21] |
KIM H, LEE K, HWANG H, BHATNAGAR N, KIM D Y, YOON I S, BYUN M O, KIM S T, JUNG K H, KIM B G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression[J]. Journal of Experimental Botany, 2014, 65(2): 453-464. DOI:10.1093/jxb/ert397 |
| [22] |
MA Y, SZOSTKIEWICZ I, KORTE A, MOES D, YANG Y, CHRISTMANN A, GRILL E. Regulators of PP2C phosphatase activity function as abscisic acid sensors[J]. Science, 2009, 324(5930): 1064-1068. DOI:10.1126/science.1172408 |
| [23] |
HAUSER F, LI Z, WAADT R, SCHROEDER J I. Snapshot: Abscisic acid signaling[J]. Cell, 2017, 171(7): 1708. DOI:10.1016/j.cell.2017.11.045 |
| [24] |
LIAO Z, ZHANG Y, YU Q, FANG W, CHEN M, LI T, LIU Y, LIU Z, CHEN L, YU S, XIA H, XUE H, YU H, LUO L. Coordination of growth and drought responses by GA-ABA signaling in rice[J]. New Phytologist, 2023, 240(3): 1149-1161. DOI:10.1111/nph.19209 |
| [25] |
HOU X, XIE K, YAO J, QI Z, XIONG L. A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(15): 6410-6415. DOI:10.1073/pnas.0901940106 |
| [26] |
JING X, SHI P, ZHANG R, ZHOU M, SHALMANI A, WANG G, LIU W, LI W, CHEN K. Rice kinase OsMRLK63 contributes to drought tolerance by regulating reactive oxygen species production[J]. Plant Physiology, 2024, 194(4): 2679-2696. DOI:10.1093/plphys/kiad684 |
| [27] |
SHI Y, CHANG Y, WU H, SHALMANI A, LIU W, LI W, XU J, CHEN K. OsRbohB-mediated ROS production plays a crucial role in drought stress tolerance of rice[J]. Plant Cell Reports, 2020, 39(12): 1767-1784. DOI:10.1007/s00299-020-02603-2 |
| [28] |
XIONG H, YU J, MIAO J, LI J, ZHANG H, WANG X, LIU P, ZHAO Y, JIANG C, YIN Z, LI Y, GUO Y, FU B, WANG W, LI Z, ALI J, LI Z. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging[J]. Plant Physiology, 2018, 178(1): 451-467. DOI:10.1104/pp.17.01492 |
| [29] |
YANG Z, WU Y, LI Y, LING H Q, CHU C. OsMTt1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice[J]. Plant Molecular Biology, 2009, 70(1/2): 219-229. DOI:10.1007/s11103-009-9466-1 |
| [30] |
PAN J, LI Z, WANG Q, YANG L, YAO F, LIU W. An S-domain receptor-like kinase, OsESG1, regulates early crown root development and drought resistance in rice[J]. Plant Science, 2020, 290: 110318. DOI:10.1016/j.plantsci.2019.110318 |
| [31] |
OUYANG S, LIU Y, LIU P, LEI G, HE S, MA B, ZHANG W, ZHANG J, CHEN S. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants[J]. Plant Journal, 2010, 62(2): 316-329. DOI:10.1111/j.1365-313X.2010.04146.x |
| [32] |
LI Y, HAN S, SUN X, KHAN N U, ZHONG Q, ZHANG Z, ZHANG H, MING F, LI Z, LI J. Variations in OsSPL10 confer drought tolerance by directly regulating OsNAC2 expression and ROS production in rice[J]. Journal of Integrative Plant Biology, 2023, 65(4): 918-933. DOI:10.1111/jipb.13414 |
| [33] |
CHEN F, ZHANG H, LI H, LIAN L, WEI Y, LIN Y, WANG L, HE W, CAI Q, XIE H, ZHANG H, ZHANG J. IPA1 improves drought tolerance by activating SNAC1 in rice[J]. BMC Plant Biology, 2023, 23(1): 55. DOI:10.1186/s12870-023-04062-9 |
| [34] |
ZHANG H, ZHAI N, MA X, ZHOU H, CUI Y, WANG C, XU G. Overexpression of OsRLCK241 confers enhanced salt and drought tolerance in transgenic rice (Oryza sativa L.)[J]. Gene, 2021, 768: 145278. DOI:10.1016/j.gene.2020.145278 |
| [35] |
JING X, LI W, ZHOU M, SHI P, ZHANG R, SHALMANI A, MUHAMMAD I, WANG G, LIU W, CHEN K. Rice carbohydrate-binding malectin-like protein, OsCBM1, contributes to drought-stress tolerance by participating in NADPH oxidase-mediated ROS production[J]. Rice, 2021, 14(1): 100. DOI:10.1186/s12284-021-00541-5 |
| [36] |
马孝松, 曾贤军, 李恩熙, 梅捍卫, 罗利军, 刘鸿艳. 水稻耐旱性及其研究进展[J]. 上海农业学报, 2022, 38(4): 36-45. DOI:10.15955/j.issn1000-3924 MA X S, ZENG X J, LI E X, MEI H W, LUO L J, LIU H Y. Advances of drought tolerance researches in rice[J]. Acta Agriculturae Shanghai, 2022, 38(4): 36-45. DOI:10.15955/j.issn1000-3924 |
| [37] |
GENG A, LIAN W, WANG Y, LIU M, ZHANG Y, WANG X, CHEN G. Molecular mechanisms and regulatory pathways underlying drought stress response in rice[J]. International Journal of Molecular Sciences, 2024, 25(2): 1185. DOI:10.3390/ijms25021185 |
| [38] |
CHEN G, LIU C, GAO Z, ZHANG Y, JIANG H, ZHU L, REN D, YU L, XU G, QIAN Q. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice[J]. Frontiers in Plant Science, 2017, 8: 1885. DOI:10.3389/fpls.2017.01885 |
| [39] |
AHMAD I, MIAN A, MAATHUIS F J. Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance[J]. Journal of Experimental Botany, 2016, 67(9): 2689-2698. DOI:10.1093/jxb/erw103 |
| [40] |
AHMAD I, DEVONSHIRE J, MOHAMED, SCHULTZE M, MAATHUIS F J. Overexpression of the potassium channel TPKb in small vacuoles confers osmotic and drought tolerance to rice[J]. New Phytologist, 2016, 209(3): 1040-1048. DOI:10.1111/nph.13708 |
| [41] |
LIU K, WANG L, XU Y, CHEN N, MA Q, LI F, CHONG K. Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice[J]. Planta, 2007, 226(4): 1007-1016. DOI:10.1007/s00425-007-0548-5 |
| [42] |
LEE D K, JUNG H, JANG G, JEONG J S, KIM Y S, HA S H, DO C Y, KIM J K. Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance[J]. Plant Physiology, 2016, 172(1): 575-588. DOI:10.1104/pp.16.00379 |
| [43] |
赵慧. 水稻耐旱性调控网络关键调节基因的克隆及OsERF65的功能研究[D]. 武汉: 华中农业大学, 2020. DOI: 10.27158/d.cnki.ghznu.2020.000448. ZHAO H. Cloning of key regulatory genes in rice drought tolerance regulatory network and functional study of OsERF65[D]. Wuhan: Huazhong Agricultural University, 2020. DOI: 10.27158/d.cnki.ghznu.2020.000448. |
| [44] |
ISLAM M A, DU H, NING J, YE H, XIONG L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance[J]. Plant Molecular Biology, 2009, 70(4): 443-456. DOI:10.1007/s11103-009-9483-0 |
| [45] |
WANG Z, TIAN X, ZHAO Q, LIU Z, LI X, REN Y, TANG J, FANG J, XU Q, BU Q. The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice[J]. The Plant Cell, 2018, 30(1): 228-244. DOI:10.1105/tpc.17.00823 |
| [46] |
JIAN L, KANG K, CHOI Y, SUH M C, PAEK N C. Mutation of OsMYB60 reduces rice resilience to drought stress by attenuating cuticular wax biosynthesis[J]. Plant Journal, 2022, 112(2): 339-351. DOI:10.1111/tpj.15947 |
| [47] |
ZHU X, XIONG L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(44): 17790-17795. DOI:10.1073/pnas.1316412110 |
| [48] |
GAO J, ZHAO Y, ZHAO Z, LIU W, JIANG C, LI J, ZHANG Z, ZHANG H, ZHANG Y, WANG X, SUN X, LI Z. RRS1 shapes robust root system to enhance drought resistance in rice[J]. New Phytologist, 2023, 238(3): 1146-1162. DOI:10.1111/nph.18775 |
| [49] |
UGA Y, SUGIMOTO K, OGAWA S, RANE J, ISHITANI M, HARA N, KITOMI Y, INUKAI Y, ONO K, KANNO N, INOUE H, TAKEHISA H, MOTOYAMA R, NAGAMURA Y, WU J, MATSOMOTO T, TAKAI T, OKUNO K, YANO M. Control of root system architecture by deeper rooting 1 increases rice yield under drought conditions[J]. Nature Genetics, 2013, 45(9): 1097-1102. DOI:10.1038/ng.2725 |
| [50] |
CHENG S, ZHOU D, ZHAO Y. WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development[J]. Plant Signaling and Behavior, 2016, 11(2): e1130198. DOI:10.1080/15592324.2015.1130198 |
| [51] |
CHAMPOUX M C, WANG G, SARKARUNG S, MACKILL D J, O'TOOLE J C, HUANG N, MCCOUCH S R. Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers[J]. Theoretical and Applied Genetics, 1995, 90(7/8): 969-981. DOI:10.1007/BF00222910 |
| [52] |
刘强明, 唐永群, 肖人鹏, 张现伟, 李经勇. 水稻耐旱的分子研究进展[J]. 分子植物育种, 2019, 17(9): 2841-2849. DOI:10.13271/j.mpb.017.002841 LIU Q M, TANG Y Q, XIAO R P, ZHANG X W, LI J Y. Progress on molecular research of drought resistance in rice[J]. Molecular Plant Breeding, 2019, 17(9): 2841-2849. DOI:10.13271/j.mpb.017.002841 |
| [53] |
SATRIO R D, FENDIYANTO M H, SUPENA E, SUHARSONO S, MIFTAHUDIN M. Genome-wide SNP discovery, linkage mapping, and analysis of QTL for morpho-physiological traits in rice during vegetative stage under drought stress[J]. Physiology and Molecular Biology of Plants, 2021, 27(11): 2635-2650. DOI:10.1007/s12298-021-01095-y |
| [54] |
BHATTARAI U, SUBUDHI P K. Genetic analysis of yield and agronomic traits under reproductive-stage drought stress in rice using a high-resolution linkage map[J]. Gene, 2018, 669: 69-76. DOI:10.1016/j.gene.2018.05.086 |
| [55] |
BARIK S R, PANDIT E, MOHANTY S P, NAYAK D K, PRADHAN S K. Genetic mapping of physiological traits associated with terminal stage drought tolerance in rice[J]. BMC Genetics, 2020, 21(1): 76. DOI:10.1186/s12863-020-00883-x |
| [56] |
SUN X, XIONG H, JIANG C, ZHANG D, YANG Z, HUANG Y, ZHU W, MA S, DUAN J, WANG X, LIU W, GUO H, LI G, QI J, LIANG C, ZHANG Z, LI J, ZHANG H, HAN L, ZHOU Y, PENG Y, LI Z. Natural variation of DROT1 confers drought adaptation in upland rice[J]. Nature Communications, 2022, 13(1): 4265. DOI:10.1038/s41467-022-31844-w |
| [57] |
YUAN X, WANG H, CAI J, BI Y, LI D, SONG F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response[J]. BMC Plant Biology, 2019, 19(1): 1-19. DOI:10.1186/s12870-019-1883-y |
| [58] |
GUO C, CHEN L, CUI Y, TANG M, GUO Y, YI Y, LI Y, LIU L, CHEN L. RNA binding protein OsTZF7 traffics between the nucleus and processing bodies/stress granules and positively regulates drought stress in rice[J]. Frontiers in Plant Science, 2022, 13: 802337. DOI:10.3389/fpls.2022.802337 |
| [59] |
MIAO J, LI X, LI X, TAN W, YOU A, WU S, TAO Y, CHEN C, WANG J, ZHANG D, GONG Z, YI C, YANG Z, GU M, LIANG G, ZHOU Y. OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice[J]. New Phytologist, 2020, 227(5): 1417-1433. DOI:10.1111/nph.16670 |
| [60] |
ZHANG J, ZHANG H, SRIVASTAVA A K, PAN Y, BAI J, FANG J, SHI H, ZHU J K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development[J]. Plant Physiology, 2018, 176(3): 2082-2094. DOI:10.1104/pp.17.01432 |
| [61] |
LI C, SHEN H, WANG T, WANG X. ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa[J]. Plant and Cell Physiology, 2015, 56(12): 2396-2408. DOI:10.1093/pcp/pcv154 |
| [62] |
ZHANG G, HOU X, WANG L, XU J, CHEN J, FU X, SHEN N, NIAN J, JIANG Z, HU J, ZHU L, RAO Y, SHI Y, REN D, DONG G, GAO Z, GUO L, QIAN Q, LUAN S. Photo-sensitive leaf rolling 1 encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice[J]. New Phytologist, 2021, 229(2): 890-901. DOI:10.1111/nph.16899 |
| [63] |
SUN L, ZHANG Q, WU J, ZHANG L, JIAO X, ZHANG S, ZHANG Z, SUN D, LU T, SUN Y. Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice[J]. Plant Physiology, 2014, 165(1): 335-345. DOI:10.1104/pp.113.232629 |
| [64] |
CHEN S, XU K, KONG D, WU L, CHEN Q, MA X, MA S, LI T, XIE Q, LIU H, LUO L. Ubiquitin ligase OsRINGzf1 regulates drought resistance by controlling the turnover of OsPIP2;1[J]. Plant Biotechnology Journal, 2022, 20(9): 1743-1755. DOI:10.1111/pbi.13857 |
| [65] |
JUNG S E, BANG S W, KIM S H, SEO J S, YOON H B, KIM Y S, KIM J K. Overexpression of OsERF83, a vascular tissue-specific transcription factor gene, confers drought tolerance in rice[J]. International Journal of Molecular Sciences, 2021, 22(14): 7656. DOI:10.3390/ijms22147656 |
| [66] |
徐靖, 唐清杰, 朱红林, 严小微, 唐力琼, 孟卫东. 水稻(Oryza sativa L.) 干旱胁迫响应转录因子研究进展[J]. 基因组学与应用生物学, 2015, 34(11): 2525-2531. DOI:10.13417/j.gab.034.002525 XU J, TANG Q J, ZHU H L, YAN X W, TANG L Q, MENG W D. Research advance of transcription factors related to rice (Oryza sativa L.) drought stress response[J]. Genomics and Applied Biology, 2015, 34(11): 2525-2531. DOI:10.13417/j.gab.034.002525 |
| [67] |
JUNG S E, KIM T H, SHIM J S, BANG S W, BIN Y H, OH S H, KIM Y S, OH S J, SEO J S, KIM J K. Rice NAC17 transcription factor en ha nces d roug ht tolera nce by modu lat ing lig nin accumulation[J]. Plant Science, 2022, 323: 111404. DOI:10.1016/j.plantsci.2022.111404 |
| [68] |
JEONG J S, KIM Y S, REDILLAS M C, JANG G, JUNG H, BANG S W, CHOI Y D, HA S H, REUZEAU C, KIM J K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field[J]. Plant Biotechnology Journal, 2013, 11(1): 101-114. DOI:10.1111/pbi.12011 |
| [69] |
LEE D K, CHUNG P J, JEONG J S, JANG G, BANG S W, JUNG H, KIM Y S, HA S H, CHOI Y D, KIM J K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance[J]. Plant Biotechnology Journal, 2017, 15(6): 754-764. DOI:10.1111/pbi.12673 |
| [70] |
JEONG J S, KIM Y S, BAEK K H, JUNG H, HA S H, DO C Y, KIM M, REUZEAU C, KIM J K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions[J]. Plant Physiology, 2010, 153(1): 185-197. DOI:10.1104/pp.110.154773 |
| [71] |
SHIM J S, OH N, CHUNG P J, KIM Y S, CHOI Y D, KIM J K. Overexpression of OsNAC14 improves drought tolerance in rice[J]. Frontiers in Plant Science, 2018, 9: 310. DOI:10.3389/fpls.2018.00310 |
| [72] |
HUANG L, HONG Y, ZHANG H, LI D, SONG F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance[J]. BMC Plant Biology, 2016, 16(1): 203. DOI:10.1186/s12870-016-0897-y |
| [73] |
EULGEM T, RUSHTON P J, ROBATZEK S, SOMSSICH I E. The WRKY superfamily of plant transcription factors[J]. Trends in Plant Science, 2000, 5(5): 199-206. DOI:10.1016/s1360-1385(00)01600-9 |
| [74] |
CHUJO T, TAKAI R, AKIMOTO-TOMIYAMA C, ANDO S, MINAMI E, NAGAMURA Y, KAKU H, SHIBUYA N, YASUDA M, NAKASHITA H, UMEMURA K, OKADA A, OKADA K, NOJIRI H, YAMANE H. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice[J]. Biochim Biophys Acta, 2007, 1769(7/8): 497-505. DOI:10.1016/j.bbaexp.2007.04.006 |
| [75] |
SONG G, SON S, LEE K S, PARK Y J, SUH E J, LEE S I, PARK S R. OsWRKY114 negatively regulates drought tolerance by restricting stomatal closure in rice[J]. Plants (Basel), 2022, 11(15): 1938. DOI:10.3390/plants11151938 |
| [76] |
SHARONI A M, NURUZZAMAN M, SATOH K, SHIMIZU T, KONDOH H, SASAYA T, CHOI I R, OMURA T, KIKUCHI S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice[J]. Plant and Cell Physiology, 2011, 52(2): 344-360. DOI:10.1093/pcp/pcq196 |
| [77] |
RASHID M, GUANGYUAN H, GUANGXIAO Y, HUSSAIN J, XU Y. AP2/ERF transcription factor in rice: Genome-wide canvas and syntenic relationships between monocots and eudicots[J]. Evolutionary Bioinformatics, 2012, 8: 321-355. DOI:10.4137/EBO.S9369 |
| [78] |
PARK S, KWON H J, CHO M H, SONG J S, KIM B, BAEK J, KIM S L, JI H, KWON T, KIM K, YOON I S. The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants[J]. International Journal of Molecular Sciences, 2021, 22(13): 7181. DOI:10.3390/ijms22137181 |
| [79] |
JUNG H, CHUNG P J, PARK S H, REDILLAS M, KIM Y S, SUH J W, KIM J K. Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance[J]. Plant Biotechnology Journal, 2017, 15(10): 1295-1308. DOI:10.1111/pbi.12716 |
| [80] |
LEE D K, YOON S, KIM Y S, KIM J K. Rice OsER F71-mediated root modification affects shoot drought tolerance[J]. Plant Signaling and Behavior, 2017, 12(1): e1268311. DOI:10.1080/15592324.2016.1268311 |
| [81] |
AHN H, JUNG I, SHIN S J, PARK J, RHEE S, KIM J K, JUNG W, KWON H B, KIM S. Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice[J]. Frontiers in Plant Science, 2017, 8: 1044. DOI:10.3389/fpls.2017.01044 |
| [82] |
ZHANG Y, LI J, CHEN S, MA X, WEI H, CHEN C, GAO N, ZOU Y, KONG D, LI T, LIU Z, YU S, LUO L. An APETALA2/ethylene responsive factor, OsEBP89 knockout enhances adaptation to direct-seeding on wet land and tolerance to drought stress in rice[J]. Molecular Genetics and Genomics, 2020, 295(4): 941-956. DOI:10.1007/s00438-020-01669-7 |
| [83] |
JIN Y, PAN W, ZHENG X, CHENG X, LIU M, MA H, GE X. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues[J]. Plant Molecular Biology, 2018, 98(1/2): 51-65. DOI:10.1007/s11103-018-0762-5 |
| [84] |
BUTT H I, YANG Z, GONG Q, CHEN E, WANG X, ZHAO G, GE X, ZHANG X, LI F. GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance[J]. BMC Plant Biology, 2017, 17(1): 142. DOI:10.1186/s12870-017-1078-3 |
| [85] |
段俊枝, 李莹, 冯丽丽, 孙岩, 齐红志, 齐学礼, 杨翠苹, 王楠, 燕照玲, 陈海燕, 张会芳, 卓文飞, 平西栓. MYB转录因子在水稻抗逆基因工程中的应用进展[J]. 江苏农业科学, 2021, 49(21): 46-53. DOI:10.15889/j.issn.1002-1302.2021.21.007 DUAN J Z, LI Y, FENG L L, SUN Y, QI H Z, QI X L, YANG C P, WANG N, YAN Z L, CHEN H Y, ZHANG H F, ZHUO W F, PING X S. Research progress on application of MYB transcription factors in rice stress tolerance genetic engineering[J]. Jiangsu Agricultural Sciences, 2021, 49(21): 46-53. DOI:10.15889/j.issn.1002-1302.2021.21.007 |
| [86] |
TANG Y, BAO X, ZHI Y, WU Q, GUO Y, YIN X, ZENG L, LI J, ZHANG J, HE W, LIU W, WANG Q, JIA C, LI Z, LIU K. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice[J]. Frontiers in Plant Science, 2019, 10: 168. DOI:10.3389/fpls.2019.00168 |
| [87] |
彭运鹏, 杨诗勤, 徐凯, 罗利军. 水稻耐旱性基因OsMYB105的表达特征和转录自激活性分析[J/OL]. 分子植物育种, 1-11[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20220411.0008.002.html. PENG Y P, YANG S Q, XU K, LUO L J. Expression patterns and transcriptional autoactivation analysis of rice drought tolerance gene OsMYB105[J/OL]. Molecular Plant Breeding, 1-11[2024-05-17]. http://kns.cnki.net/kcms/detail/46.1068.S.20220411.0008.002.html. |
| [88] |
FAVERO P R, MARA D A L, DOS S B M, MACEDO N P, PALMA B M A, DOMINGUES C S, VASCONCELOS R R, DE SOUZA G M, NEBO C, VARGAS D, CRESTE S. Overexpression of ScMYBAS1 alternative splicing transcripts differentially impacts biomass accumulation and drought tolerance in rice transgenic plants[J]. PloS One, 2018, 13(12): e207534. DOI:10.1371/journal.pone.0207534 |
| [89] |
YIN X, CUI Y, WANG M, XIA X. Overexpression of a novel MYB-related transcription factor, OsMYBR1, confers improved drought tolerance and decreased ABA sensitivity in rice[J]. Biochemical and Biophysical Research Communications, 2017, 490(4): 1355-1361. DOI:10.1016/j.bbrc.2017.07.029 |
| [90] |
XIONG H, LI J, LIU P, DUAN J, ZHAO Y, GUO X, LI Y, ZHANG H, ALI J, LI Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice[J]. PLoS One, 2014, 9(3): e92913. DOI:10.1371/journal.pone.0092913 |
| [91] |
XU W, TANG W, WANG C, GE L, SUN J, QI X, HE Z, ZHOU Y, CHEN J, XU Z, MA Y Z, CHEN M. SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway[J]. Frontiers in Plant Science, 2020, 11: 785. DOI:10.3389/fpls.2020.00785 |
| [92] |
YANG L, CHEN Y, XU L, WANG J, QI H, GUO J, ZHANG L, SHEN J, WANG H, ZHANG F, XIE L, ZHU W, LYU P, QIAN Q, YU H, SONG S. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice[J]. Molecular Plant, 2022, 15(7): 1227-1242. DOI:10.1016/j.molp.2022.06.003 |
| [93] |
QU X, ZOU J, WANG J, YANG K, WANG X, LE J. A rice R2R3-type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC[J]. International Journal of Molecular Sciences, 2022, 23(11): 5873. DOI:10.3390/ijms23115873 |
| [94] |
仝宇, 王聪, 赵利利, 连娟, 刘晓梅, 赵宝存. 转录因子OsbZIP5负调控水稻的耐旱性[J]. 中国生物化学与分子生物学报, 2021, 37(6): 798-810. DOI:10.13865/j.cnki.cjbmb.2021.03.1427 TONG Y, WANG C, ZHAO L L, LIAN J, LIU X M, ZHAO B C. Transcription factor OsbZIP5 negative regulates drought-tolerance in rice[J]. Chinese Journal of Biochemistry and Molecular Biology, 2021, 37(6): 798-810. DOI:10.13865/j.cnki.cjbmb.2021.03.1427 |
| [95] |
杨诗勤. 水稻耐旱转录因子基因的分离和OsbZIP62的功能鉴定[D]. 武汉: 华中农业大学, 2019. DOI: 10.27158/d.cnki.ghznu.2019.000963. YANG S Q. Isolation of drought tolerance related transcription factor gene and functional characterization of OsbZIP62 in rice[D]. Wuhan: Huazhong Agricultural University, 2019. DOI: 10.27158/d.cnki.ghznu.2019.000963. |
| [96] |
SHENG P, TAN J, JIN M, WU F, ZHOU K, MA W, HENG Y, WANG J, GUO X, ZHANG X, CHENG Z, LIU L, WANG C, LIU X, WAN J. Albino midrib 1, encoding a putative potassium efflux antiporter, affects chloroplast development and drought tolerance in rice[J]. Plant Cell Reports, 2014, 33(9): 1581-1594. DOI:10.1007/s00299-014-1639-y |
| [97] |
GUO C, GE X, MA H. The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages[J]. Plant Molecular Biology, 2013, 82(3): 239-253. DOI:10.1007/s11103-013-0057-9 |
| [98] |
LIU S, ZHENG L, XUE Y, ZHANG Q, WANG L, SHOU H. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice[J]. Journal of Plant Biology, 2010, 53(6): 444-452. DOI:10.1007/s12374-010-9135-6 |
| [99] |
BAE H, KIM S K, CHO S K, KANG B G, KIM W T. Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.)[J]. Plant Science, 2011, 180(6): 775-782. DOI:10.1016/j.plantsci.2011.02.008 |
| [100] |
KIM S, PARK S I, KWON H, CHO M H, KIM B G, CHUNG J H, NAM M H, SONG J S, KIM K H, YOON I S. The rice abscisic acid-responsive RING finger E3 ligase OsRF1 targets OsPP2C09 for degradation and confers drought and salinity tolerance in rice[J]. Frontiers in Plant Science, 2021, 12: 797940. DOI:10.3389/fpls.2021.797940 |
| [101] |
DU H, WU N, CUI F, YOU L, LI X, XIONG L. A homolog of ethylene overproducer, OsETOL1, differentially modulates drought and submergence tolerance in rice[J]. Plant Journal, 2014, 78(5): 834-849. DOI:10.1111/tpj.12508 |
| [102] |
LYU Q, LI X, JIN X, SUN Y, WU Y, WANG W, HUANG J. Rice OsPUB16 modulates the 'SAPK9-OsMADS23-OsAOC' pathway to reduce plant water-deficit tolerance by repressing ABA and JA biosynthesis[J]. Plos Genetics, 2022, 18(11): e1010520. DOI:10.1371/journal.pgen.1010520 |
| [103] |
CUI L H, MIN H J, BYUN M Y, OH H G, KIM W T. OsDIRP1, a putative RING E3 ligase, plays an opposite role in drought and cold stress responses as a negative and positive factor, respectively, in rice (Oryza sativa L.)[J]. Frontiers in Plant Science, 2018, 9: 1797. DOI:10.3389/fpls.2018.01797 |
| [104] |
PARK Y C, CHOI S Y, KIM J H, JANG C S. Molecular functions of rice cytosol-localized RING finger protein 1 in response to salt and drought and comparative analysis of its grass orthologs[J]. Plant and Cell Physiology, 2019, 60(11): 2394-2409. DOI:10.1093/pcp/pcz133 |
| [105] |
NING J, LI X, HICKS K M, XIONG L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice[J]. Plant Physiology, 2010, 152(2): 876-890. DOI:10.1104/pp.109.149856 |
| [106] |
YOU J, ZONG W, HU H, LI X, XIAO J, XIONG L. A Stress-responsive NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice[J]. Plant Physiology, 2014, 166(4): 2100-2114. DOI:10.1104/pp.114.251116 |
| [107] |
LIU B, FAN J, ZHANG Y, MU P, WANG P, SU J, LAI H, LI S, FENG D, WANG J, WANG H. OsPFA-DSP1, a rice protein tyrosine phosphatase, negatively regulates drought stress responses in transgenic tobacco and rice plants[J]. Plant Cell Reports, 2012, 31(6): 1021-1032. DOI:10.1007/s00299-011-1220-x |
| [108] |
陆展华, 刘维, 王石光, 巫浩翔, 王晓飞, 陈浩, 潘朝阳, 何秀英. 国内外旱稻研究现状及对广东的启示[J]. 广东农业科学, 2023, 50(12): 62-72. DOI:10.16768/j.issn.1004-874X.2023.12.006 LU Z H, LIU W, WANG S G, WU H X, WANG X F, CHEN H, PAN C Y, HE X Y. Research status of upland rice worldwide and its enlightenment to Guangdong Province[J]. Guangdong Agricultural Sciences, 2023, 50(12): 62-72. DOI:10.16768/j.issn.1004-874X.2023.12.006 |
| [109] |
潘大建, 李晨, 范芝兰, 孙炳蕊, 陈文丰, 江立群, 张静, 吕树伟, 刘清, 毛兴学. 广东省农业科学院水稻种质资源研究60年: 成就与展望[J]. 广东农业科学, 2020, 47(11): 18-31. DOI:10.16768/j.issn.1004-874X.2020.11.003 PAN D J, LI C, FAN Z L, SUN B R, CHEN W F, JIANG L Q, ZHANG J, LYU S W, LIU Q, MAO X X. Sixty years' researches on rice germplasm resources of guangdong academy of agricultural sciences: Achievements and prospects[J]. Guangdong Agricultural Sciences, 2020, 47(11): 18-31. DOI:10.16768/j.issn.1004-874X.2020.11.003 |
| [110] |
潘大建, 范芝兰, 邹建运. 广东野生稻种质资源保护与育种利用[J]. 广东农业科学, 2022, 49(9): 92-104. DOI:10.16768/j.issn.1004-874X.2022.09.010 PAN D J, FAN Z L, ZOU J Y. Conservation and breeding utilization of wild rice germplasm resources in Guangdong Province[J]. Guangdong Agricultural Sciences, 2022, 49(9): 92-104. DOI:10.16768/j.issn.1004-874X.2022.09.010 |
(责任编辑 马春敏)
 2024, Vol. 51
2024, Vol. 51