文章信息
基金项目
- 国家自然科学基金(32072580)
作者简介
-
雷建军,博士,华南农业大学园艺学院二级教授,博士生导师,国务院政府特殊津贴专家,农业农村部有突出贡献的中青年专家,南粤优秀教师。在华南农业大学退休后现被韶关学院聘请指导学科建设。曾担任全国高等农业院校教学指导委员会园艺学科组副组长,中国园艺学会常务理事,国家植物航天育种中心副主任。长期从事蔬菜作物遗传育种与分子生物学研究。承担国家“863”计划、国家科技支撑计划、国家自然科学基金等项目多项;荣获省部级科技进步一等奖1项、二等奖2项,省部级教学成果一等奖1项;发表学术论文250多篇。在十字花科蔬菜作物、辣椒常规育种和蔬菜基因工程研究方面取得多项成果;选育出一系列辣椒和十字花科蔬菜新品种;阐明了辣椒素在数量上的差异的本质原因,揭示了调控辣椒素生物合成的分子机理,该结果属于国际领先;在蔬菜作物转基因研究方面获得抗虫、抗病、雄性不育、抗逆、延缓衰老等一系列转基因材料。
雷建军(1957—),男,博士,教授,研究方向为蔬菜育种与分子生物学,E-mail:jjlei@scau.edu.cn.
文章历史
- 收稿日期:2023-12-21
2. 华南农业大学园艺学院/农业农村部华南地区园艺作物生物学与种质创制重点实验室,广东 广州 510642
2. College of Horticulture, South China Agricultural University/Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (South China), Ministry of Agriculture and Rural Affairs, Guangzhou 510642, China
辣椒(Capsicum annuum L.)起源于南美洲,16世纪末传到中国[1]。由于我国地域广阔、生态类型多样,辣椒在如此多样的环境条件下经过长时间的自然驯化和品种改良,产生很多类型,如灯笼椒、牛角椒、朝天椒、线椒、炮椒、螺丝牛角椒、螺丝线椒、海南黄灯笼、涮涮辣和小米辣等。辣度差异很大,有不辣的甜椒,也有很辣的海南黄灯笼和涮涮辣,可以满足不同消费人群的需求。辣椒的营养价值高,在保健、美容、食品、医药、制暴等行业均有重要作用,已成为我国最重要的蔬菜作物之一[2],栽培面积逐渐增加,目前已经位于我国蔬菜作物之首,据国家大宗蔬菜产业技术体系统计,辣椒年种植面积达到213万hm2[3]。辣椒在栽培过程中,由于多方面原因而遭受各种病害(病毒病、青枯病、疫病、炭疽病、疮痂病等)的侵袭,给生产造成重大损失。通过化学防治方法、农业防治措施等可以在一定程度上减轻为害,但选用抗病品种才是防治病害最经济有效的措施。我国从第6个“五年计划”开始日益重视辣椒的抗病育种,组织全国从事辣椒育种和病理的专家联合攻关,取得显著成绩[1-6]。以往主要通过常规育种改良选育抗病品种,但仅依赖常规育种方法是不够的。随着分子生物学的发展,抗病分子育种与常规育种方法相结合是将来取得突破的发展方向。近年来,辣椒抗病分子育种取得了很多重要进展[7-8],分子育种主要包括两部分:一是分子标记辅助选择,二是利用基因工程创制抗病育种材料。在分子标记辅助选择中,首先必须找到分子标记。分子标记分为基因标记、构建分离群体进行连锁分析获得的标记以及通过大量材料测序进行关联分析获得的标记。基因工程的前提是分离获得抗病基因,然后将抗病基因导入到经济性状优良的辣椒育种材料中,或者通过基因编辑技术或基因沉默技术使负向调控的抗病基因发生突变,从而提高辣椒抗病性。本文将从分子标记及其辅助选择、抗病分子机理(青枯病、疫病、炭疽病、病毒病、疮痂病和软腐病等)、采用转基因技术创制抗病种质(抗病毒病、抗青枯病和抗疫病等)3个方面进行综合介绍,并对今后辣椒抗病育种领域的发展趋势和研究重点进行展望。
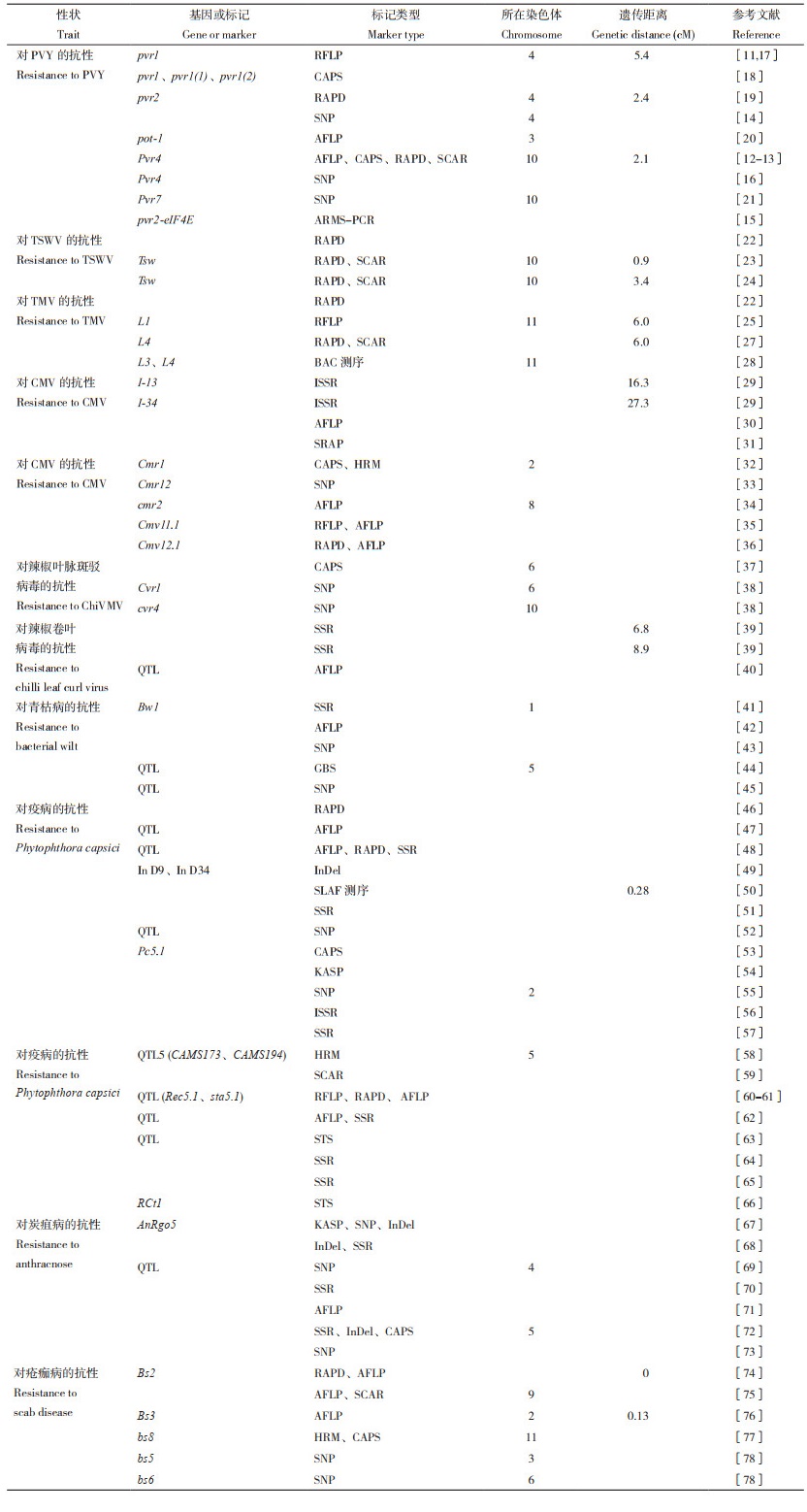
1 辣椒抗病性分子标记及其辅助选择分子标记经历了从连锁标记、基因标记(功能标记)到关联标记的发展过程。前期主要是利用分离群体寻找连锁标记,连锁标记的缺点是通用性不足,更换群体后精确性可能发生改变。基因标记是随着基因分离技术的完善而发展的,基因标记通用性很强,不受群体来源的影响,准确率极高。关联标记是随着DNA测序技术的发展而发展的,随着DNA测序技术的发展,测序成本越来越低,对大量材料进行重测序,为关联标记的筛选提供了基础。
1.1 分子标记开发分子标记开发是分子标记辅助选择的前提。早期开发的分子标记都是通过构建分离群体,按照孟德尔遗传规律进行连锁分析获得与抗病性连锁的标记。随着抗病分子机理研究的不断深入,克隆分离得到一些抗病基因,用抗病基因作分子标记直接进行选择,选择精准,效率高。随着DNA测序技术的不断完善,测序成本越来越低,对大量育种材料进行大规模测序成为可能,通过关联分析开发了很多SNP(Single nucleotide polymorphism,单核苷酸多态性)标记。目前对辣椒主要病害均开发了分子标记。
1.1.1 对PVY(Potato-virus Y,马铃薯Y病毒)的抗性 控制辣椒对PVY 抗性的基因有很多,其中pvy6[9-10]尚未找到分子标记,其他均已找到分子标记。Murphy等[11]找到与pvr1 连锁的RFLP(Restriction fragment length polymorphism,限制性片段长度多态性)标记。Caranta等[12]找到Pvr4的CAPS(Cleaved amplified polymorphic sequence,酶切扩增多态性序列)标记,离其最近的标记只有2.1 cM(表 1)。Arnedo-Andrés等[13]找到与Pvr4连锁的RAPD(Randomly amplified polymorphic DNA,随机扩增多态性DNA)和SCAR(Sequence characterised amplified region,特定序列扩增)标记。Tamisier等[14]通过基因组重测序,采用全基因组关联分析法找到多个抗PVY 基因的SNP分子标记(位于4、6、9、12号染色体),其中在4号染色体上有pvy2的分子标记。Rubio等[15]利用ARMS-PCR(Amplification refractory mutation systemPolymerase chain reaction,扩增抑制突变系统PCR)获得pvr2-eIF4E的功能标记。Devran等[16]采用新一代测序技术获得与Pvr4紧密连锁的SNP标记。Holdsworth等[17]利用KASP(Kompetitive allele-specific PCR,竞争性等位基因特异性PCR)找到pvr1 的功能标记(基因标记)。其他标记详见表 1[18-21]。
1.1.2 对TSWV(Tomato spotted wilt virus,番茄斑点萎蔫病毒)的抗性 Loaiza-Figueroa[22]找到与抗TSWV特性连锁的RAPD标记。Moury等[23]在第10号染色体上找到抗TSWV的SCAR标记,离其最近的标记只有0.9 cM、较远的为3.4 cM;抗病对感病为显性[23-24]。
1.1.3 对TMV(Tobacco mosaic virus,烟草花叶病病毒)的抗性 辣椒对TMV的抗性是由L基因控制的,L有一系列等位基因,其中L1位于一年生辣椒的第11号染色体、离番茄的RFLP标记TG36较近(6.0 cM),L4来源于C. chacoense[25]。Lefebvre等[26]找到抗TMV的RAPD和AFLP标记。Sugita等[27]找到抗TMV的RAPD标记,并已经转化成SCAR标记(1.5 cM)。Yang等[28]通过BAC(Bacerial artificial chiromsome,细菌人工染色体)克隆测序找到与抗TMV相关的L3和L4基因连锁的标记。
1.1.4 对CMV(Cucumber mosaic virus,黄瓜花叶病毒)的抗性 王述彬等[29]利用ISSR技术找到2个标记(I-13和I-34),其中I-13的遗传距离为16.3 cM,该特异性片段约为1 100 bp,命名为I-131100;I-34与辣椒抗CMV抗病位点连锁,遗传距离为27.3 cM,该特异性片段约为450 bp,命名为I-34450。杨学玲[30]找到与抗CMV特性连锁的AFLP标记(E18/M06),连锁距离为25.4 cM,该特异性片段约为450 bp。赵娟[31]构建了一张有13个连锁群的辣椒遗传图谱,共76个分子标记,其中SRAP标记55个、SSR标记21个,图谱覆盖长度为830.4 cM,标记间平均图距为10.92 cM,长度处于0~152.2 cM范围内;并利用MapQTL4.0软件中的复合区间作图法对辣椒抗CMV进行QTL分析,结果在第1、4、7连锁群上分别检测到1个QTL位点,其中与第1连锁群上的QTL位点距离最近的两个标记是C9C10-4、B11C16-2,与第4连锁群上的QTL位点距离最近的标记是B13C10、HpmsE072,与第7连锁群上的QTL位点距离最近的标记是HpmsE124、hpms2-2,这3个QTL解释表型差异的贡献率分别为12.7%、38.8% 和11.0%。其他标记详见文献[32-36]。
1.1.5 对辣椒叶脉斑驳病毒的抗性 Lee等[37]找到抗辣椒叶脉斑驳病毒的CAPS分子标记。Lee等[38]找到2个抗病基因(显性抗病基因Cvr1和隐形抗病基因cvr4)的分子标记并定位在第6和第10号染色体上。
1.1.6 对辣椒卷叶病毒的抗性 Thakur等[39]找到抗辣椒卷叶病毒的SSR标记。Dwivedi等[40]利用AFLP标记对辣椒卷叶病毒的抗性进行QTL分析,找到2个主效QTL。
1.1.7 对青枯病的抗性 Mimura等[41]开发了与抗青枯病基因连锁的标记(CAMS 451)。Lee等[44]报道了辣椒对2个青枯病菌分离物抗性的QTL,分别找到5个QTL(Bwr6w-7.2、Bwr6w-8.1、Bwr6w-9.1、Bwr6w-9.2和Bwr6w-10.1)和3个QTL(Bwr6w-5.1、Bwr6w-6.1、Bwr6w-7.1)。Chae等[45]利用基因测序分型法,获得1 550个抗青枯病的SNP标记,并构建了1个图谱。其他报道见表 1[42-43]。
1.1.8 对疫病的抗性 马海滨[46]找到1个与辣椒抗疫病特性连锁的RAPD标记。易图永[47]获得几个与辣椒抗疫病性状相关的位点QTL,可以解释表型差异的贡献率达64% 以上。安静[48]结合该课题组之前的RAPD和SSR标记,利用AFLP标记在第4连锁群上检测到2个QTL位点,与其中一个QTL位点距离最近的标记(E41M53-7)仅0.4 cM;与另一个QTL位点距离最近的分子标记(E41M52-6)为7.0 cM,两个QTL解释表型差异的贡献率分别为9.5% 和16.4%。李儒剑[49]获得2个对疫病抗感性鉴定准确率分别为78.26% 和79.31% 的标记In D9和In D34。Li等[50]通过SLAF(Specific-locus amplified fragment sequencing,特异位点扩增片段)测序与QTL分析方法构建了辣椒抗疫病的高密度遗传图谱。Kumar等[51]找到1个抗疫霉菌印度分离物的分子标记。Lozada等[52]利用SNP标记,找到抗疫病基因富集区,鉴定了一部分与抗病基因紧密连锁的候选基因。Bongiorno等[53]开发了与抗疫病基因Pc5.1连锁的CAPS(Cleaved amplified polymorphic sequence)标记。Zhang等[54]利用全基因组关联分析获得30个KASP(Kompetitive allele-specific PCR)标记。Ro等[55]利用全基因组关联分析找到多个与抗疫病特性相关的SNP标记,这些标记分布在第5、9和11号染色体以外的染色体上,其中2号染色体上的标记“Chr02-1126”的准确率为78.5%。Mohammadbagheri等[56]找到3个与抗疫病特性紧密连锁的ISSR标记。Bukhuri等[57]找到2个与抗疫病特性紧密连锁的SSR标记CAMS173和CAMS194。其他报道详见表 1[58-61]。
1.1.9 对炭疽病的抗性 Lee等[62]利用SSR标记鉴定了辣椒对炭疽病抗性的QTL,随后陆续有不少报道(表 1)[63-71]。Chen等[70]找到抗炭疽病的SSR标记。Ro等[73]利用全基因组关联分析找到25个基于田间试验结果的与抗炭疽病相关的SNP标记,以及32个基于室内接种鉴定结果的SNP标记。
1.1.10 对疮痂病的抗性 疮痂病病原菌有7个生理小种,每个小种都有一个对应的抗病基因,如Bs1抗第0、2、5小种,Bs2抗第0、3小种,Bs3抗第0、1、4小种,目前尚未找到抗第6小种的材料。Tai等[74]利用野生辣椒(C. chacoense)作抗源,与一年生辣椒杂交得到近等基因系后进行分子标记,得到与抗病基因共分离的AFLP标记(A2)。Truong等[75]也得到相似结果。Pierre等[76]找到与Bs3相连锁的分子标记,相距1.0 cM。Sharma等[77]将与bs8连锁的标记定位在11号染色体上。Sharma等[78]将小种非特异性的隐性抗病基因bs5和bs6分别定位在第3号染色体和第6号染色体上。
1.2 辅助选择利用基因标记进行选择的效率最高,而且不受群体来源的影响,因此应尽可能利用基因标记进行选择。目前分离获得的抗青枯病基因相对较多,但总体来说数量有限,在没有基因标记可以利用时,只能利用与抗病基因紧密连锁的标记进行辅助选择。
王立浩等[79]利用AFLP、SCAR和CAPS标记对马铃薯Y病毒的抗性进行选择,从中筛选出12个主效QTL上纯合抗性位点的单株。Moodley等[80]选育了一个抗PVY的辣椒品系。于海龙等[81]利用PMMoV(Pepper mild mottle virus,辣椒轻斑驳病毒)抗病位点连锁标记IH1-04、087H3T7分别检测出6份和8份辣椒种质携带目标标记,人工接种鉴定结果与标记087H3T7一致,8份辣椒种质均表现抗病;4份辣椒种质携带TSWV抗病基因Tsw连锁标记SCAC568片段,与人工接种鉴定结果相符;10份辣椒种质经疮痂病抗性基因Bs2连锁标记Bs2-L1/R1、Bs2-S45和Bs2-14F/R检测均为阳性;采用针刺法接种炭疽菌后并未发现抗炭疽病种质。李子雄等[82]利用抗性分子标记鉴定筛选出8份材料携带L4,23份材料携带CaPhyto,1份材料携带Pvr4。陈灵芝等[83]利用与L3连锁的3个分子标记(189D23MF、PMFR11和PMFR21)对辣椒种质资源进行鉴定,结果发现只有PMFR21可以用来准确鉴定辣椒对TMV的抗性。陈建分等[84]利用分子标记鉴定了辣椒对PMMoV的抗性。Fidan等[85]利用分子标记对土耳其辣椒抗黄瓜花叶病毒特性丧失情况进行检测,发现分子标记鉴定结果与田间鉴定结果一致。Ozkaynak等[86]利用分子标记辅助选择选育出一个同时抗PVY、TSWV和PMMoV的材料。Polat等[87]利用CAPS标记辅助选择获得抗TSWV的材料。
Sood等[88]利用抗青枯病的SSR标记CAMS 451辅助筛选了一些抗病材料。
郭爽等[89]利用2个分子标记鉴定辣椒抗疫病材料,发现26份材料2个标记引物的分子标记鉴定结果与田间调查结果完全吻合,包括22份抗病材料、4份感病材料;3份材料的分子标记鉴定结果与田间调查结果相反。Ates等[90]利用SCAR标记OpD04.717辅助选择选育获得7份抗疫病材料。Rabuma等[91]利用SSR标记从233份辣椒材料中鉴定获得高抗疫病材料22份、中抗材料17份。
Kim等[92]利用富鲁达纳米流体动态芯片,同时对疫病、炭疽病、疮痂病、黄瓜花叶病毒病、辣椒轻斑驳病毒病等辣椒多种病害抗性基因进行检测筛选,发现其中20个SNP标记最为准确,可以对多种病害抗性进行准确选择。
2 辣椒抗病分子机理植物为抵抗病原菌的入侵,进化出两层先天免疫系统。第一层免疫系统是由细胞膜表面定位的模式识别受体(Pattern-recognition receptor,PRR)直接识别病原体相关分子模式(Pathogenassociated molecular pattern,PAMP),而触发植物的免疫系统(Pattern-triggered immunity,PTI)。为促进入侵和增殖,许多病原体都会产生毒力相关分子进入植物细胞或质外体,以抑制宿主免疫。植物细胞内NLR(Nucleotide-binding,Leucine-rich Repeat proteins)受体蛋白通过直接或间接感知病原菌的效应因子(Effector)从而触发第二层免疫信号,称为ETI(Effector-triggered immunity)。根据能动性划分,植物抵抗病原菌的方式可以分为2种:一种是被动抗病,如在细胞表面形成一种抗病物质,从物理上阻碍病原菌的入侵;另一种是主动抗病,在植物体内形成一些抗病物质,从化学上抵御病原菌的侵染,如形成酚类物质、不饱和内酯和抗菌肽等。在主动抗病过程中,植物被病原菌侵染后迅速释放活性氧、产生过敏反应、形成植物抗毒素、加固和修复细胞壁[93]。Ding等[93]对信号转导通路归纳如下:(1)信使信号分子介导的信号通路,包括钙离子、活性氧、一氧化氮等;(2)激素介导的信号通路,包括水杨酸、茉莉酸甲酯、乙烯等;(3)异源三聚体G蛋白介导的信号通路;(4)丝裂原活化蛋白激酶(Mitogen-activated protein kinase,MAPK)级联介导的信号通路;(5)非编码RNA介导的信号通路;(6)各种信号通路相互作用形成的信号通路。以上是从宏观上介绍植物抗病的机理,而宏观上的抗病机理都要由微观的抗病基因来执行。雷建军等[7]在《辣椒分子育种研究进展》一文中涉及一部分,下面针对辣椒主要病害介绍抗病基因的分离、功能鉴定及其抗病分子机理。
2.1 抗青枯病研究表明,CaWRKY27[94]、CaHDZ27[95]、CaLRR51[96]、CaASR1[97]可以正向调控辣椒对青枯病的抗性。CaMLO6[98]是青枯病的负调控因子(与抗逆性相反),部分受CaWRKY40的调控。CaWRKY28 Cys249复合体是CaWRKY40调控辣椒对青枯病菌免疫所必需的[99]。CaCBL1沉默后,辣椒对青枯病的抗性明显降低,在高温下更明显[100]。CaPti1-CaERF3复合体共调控辣椒对青枯病的抗性,同时也耐脱水[101]。Hussain等[102]报道,CaASHH3正向调控辣椒抗青枯病。CaCDPK29-CaWRKY27b复合体促进CaWRKY40调控的辣椒对青枯病的抗性[103]。CaWRKY40调控CabZIP23,CabZIP23调控CabZIP63,形成一个正向反馈调控环,共同调控辣椒对青枯病的抗性[104]。CaREM1.4与CaRIN4互作调控辣椒对青枯病的抗性,CaRIN4-12能抑制由CaREM1.4引起的活性氧迸发及细胞坏死,表明CaREM1.4是辣椒与青枯菌互作的正调控因子,而CaRIN4-12通过与前者相互作用并影响其功能,发挥负调控作用[105]。Yang等[106]研究发现辣椒在高温高湿条件下未接种或接种青枯雷尔氏菌后早期阶段,CaKAN3和CaHSF8会在细胞核内上调表达,并在蛋白水平上相互作用。CaKAN3和CaHSF8通过激活一部分核苷酸结合和富含亮氨酸重复结构蛋白(NLRs),即在感知未知病原体效应因子时启动免疫信号,从而协同抵御青枯雷尔氏菌的侵染。当高温高湿条件持续存在而没有病原体侵袭,或者温度进一步升高时,CaHSF8则不再与CaKAN3相互作用,而是直接上调一部分热休克蛋白基因,从而激活热耐受性。CaKAN3-CaHSF8通过不同关联介导高温特异性辣椒免疫和热耐受性。MADS盒转录因子AGL8与染色质重塑组分SWC4相互作用,激活辣椒的耐热性和环境依赖性免疫(青枯病菌)[107]。丛枝菌根(Arbuscular mycorrhizal)接种后,可以诱导水杨酸和茉莉酸信号途径的基因高水平表达[108]。
2.2 抗疫病几丁质酶蛋白CaChi和聚半乳糖酶抑制蛋白CaPGIP1可以直接抑制辣椒疫霉菌的生长[109-111],并通过减少活性氧的积累、触发超敏反应(HR)和上调防御相关基因表达水平,进而增强辣椒对疫病的抗性。乙烯反应因子CaAP2/ ERF064和CaPTI1负责触发细胞死亡,并参与茉莉酸/乙烯(JA/ET)信号通路,调节CaBPR1、CaBPR1、CaDEF1和CaSAR82的表达[112-113]。然而,SBP-box家族CaSBP08、CaSBP11和CaSBP12基因沉默植株表现出对疫霉菌的抵御能力增强,伴随着防御相关基因被强烈诱导,细胞损伤降低,表明这些基因对辣椒植株抵御疫霉菌侵染的防御反应具有负调控作用[114-116]。此外,CanPOD[109]、CaRGA2[117]、CaDIR7[118]和CaCIPK1[119]等基因也被报道可增强辣椒植株对疫病的抗性。CaWRKY08-4和CaWRKY01-10两个转录因子在抗病的CM334受疫病菌浸染后高表达,而在感病的EC01中没有变化[120]。Dof转录因子可以调控辣椒对疫病的抗性[121]。Zhang等[116]报道,CaSBP12负调控辣椒对疫病的抗性;其后来又从辣椒中分离出15个SPB转录因子基因,表明CaSBP08和CaSBP11是2个负向调控因子,沉默后抗病性明显增强,在烟草中植株超表达,病情指数明显增加[114]。CaChiVI2为辣椒的抗疫病基因,同时也抗热[111]。Du等[122]报道辣椒对疫病的抗性受表观遗传的调控。辣椒对疫病相关防卫基因(PR-1b、CaWRKY58、CPI、MIR、HMG和PAL)的表达会受到根结线虫的影响[123]。CaCP15负向调控辣椒对疫病的抗性[124]。
2.3 抗炭疽病辣椒醇(Capsidiol)是一种倍半萜类物质,其合成相关基因对辣椒炭疽病菌有明显抗性,接种病原菌后,这些基因的表达量增加50多倍,但尚未作转基因鉴定[125]。Lee等[126]从抗病辣椒中分离出抗菌蛋白基因CaAMP1,该基因可以抗多种病害(17种病原菌),其中对炭疽病的抗性研究得比较清楚。研究表明,仅存在于C. baccatum中的CbCN能够抗炭疽病[127]。CaWRKY50负向调控对炭疽病的抗性[128]。两个羧酸酯酶PepEST1和PepEST2结构相似,均能调控辣椒对炭疽病的抗性,以PepEST2控制的抗性更强[129]。类受体激酶CaRLK1、CaRLK15和CaRLK16与辣椒抗炭疽病有关[130]。CbCN和CbAR9(来自C. baccatum)能抗炭疽病[127,131]。Mishra等[132]利用基因编辑技术突变CaERF28获得对炭疽病抗性增强的植株,说明它是一个负调控因子。
2.4 抗枯萎病Lee等[126]将辣椒CaAMP1在拟南芥中超表达,发现植株对枯萎病的抗性明显增强。酸性几丁质酶3、酸性葡聚糖酶、金属硫蛋白2b和渗透蛋白样PR-5可以激活辣椒的防御反应,从而抵抗病原菌的浸染[133]。CaChi2通过分解病原菌的细胞壁,CaBglu通过激活辣椒的防御反应而抗病[134]。CaChitIV与CaPIK1互作激活辣椒的防御反应,产生坏死,从而阻止病原菌的蔓延[135]。
2.5 抗病毒病Dof转录因子可以调控辣椒对TMV和辣椒斑驳病毒的抗性[121]。Koeda等[136]通过图位克隆法获得一个隐性抗病基因pepy-1(Pepper yellow leaf curl virus抗辣椒黄叶卷曲病毒)。CaNAC1可以调控辣椒对斑驳病毒的抗性[137]。辣椒抗病品种可以通过生长素途径基因的超表达,产生过敏性坏死反应,进而抗病[138]。Luo等[139]分离得到与抗黄瓜花叶病毒相关的MicroRNA。辣椒轻斑驳病毒(PMMoV)与辣椒叶绿体外膜蛋白OMP24相互作用,能抵抗病毒浸染[140]。
2.6 抗疮痂病Wang等[141]从抗病的柔毛辣椒(C. pubescens)中分离出一个广谱性的抗疮痂病基因并导入到水稻中,提高了水稻对白叶枯病的抗性。Sendin等[142]从C. chacoense中分离出CcBs2(抗疮痂病基因)导入到柑橘中后,增强了柑橘对溃疡病的抗性。Szabo等[143]通过图位克隆法获得隐性抗病基因bs5,它比野生型显性基因Bs5减少2个氨基酸。CaWRKY40a通过与病原菌的效应子XopS互作,从而提高辣椒对疮痂病的抗性[144]。
2.7 抗软腐病Djami-Tchatchou等[145]分析辣椒接种软腐病菌前后的转录组和代谢组差异,找到与抗病性相关的结构基因和转录因子,但基因功能尚未鉴定。Ger等[146]将从辣椒中分离得到的铁氧化还原蛋白基因CaPFLP导入拟南芥,提高了拟南芥对软腐病的抗性。
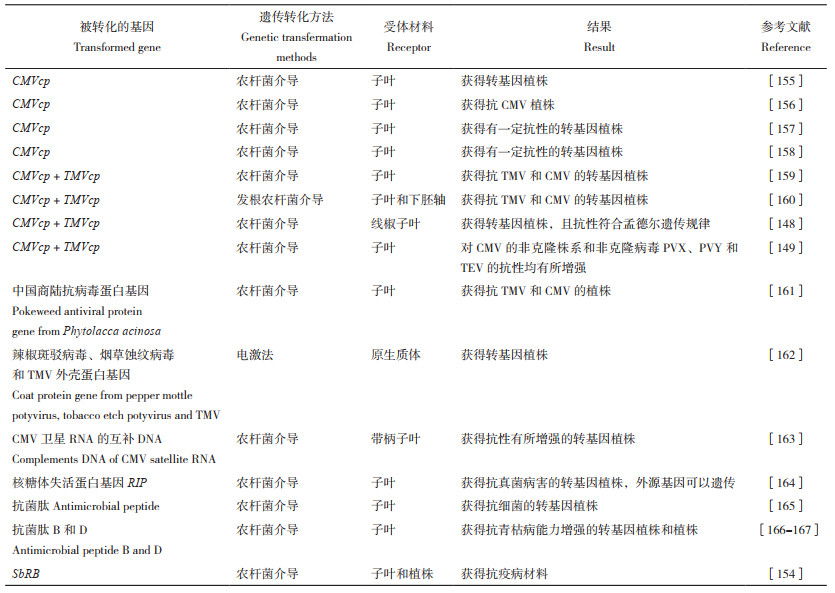
3 采用基因工程技术创制抗病辣椒种质采用常规育种方法已获得一些辣椒抗病育种原始材料,但获得的品种并不多。随着基因工程技术的发展,通过基因工程技术获得辣椒抗病品种成为重要的发展方向。早在1999年,北京大学便获得抗病毒病的转基因辣椒安全证书[147],后来又有多个转病毒外壳蛋白基因的报道[148-150]。杨国顺[151]将辣椒茉莉酸甲酯和乙烯响应结合因子基因导入辣椒,从而获得对青枯病、疫病和疮痂病抗性增强的辣椒材料。高玉尧等[152]将中国商陆抗病毒蛋白基因和苏云金杆菌晶体毒蛋白基因导入辣椒,获得抗黄瓜花叶病毒和斜纹夜蛾的材料。黄真池[153]将PFLP(Plant ferridoxin-like protein,植物类铁氧还蛋白基因)和HRAP(Hypersensitive response assisting protein,超敏反应辅助蛋白)构建双价载体在病原菌诱导型启动子启动下导入辣椒,获得抗青枯病和疫病的辣椒材料。Bagga等[154]采用农杆菌介导的渗入法和叶盘法,将来自马铃薯的抗病基因SbRB导入辣椒,获得抗疫病的转基因植株。Mishra等[132]利用基因编辑技术使CaERF28发生突变,获得抗炭疽病植株。通过转基因技术获得的辣椒抗病种质的情况详见表 2。
|
4 结语与展望
随着分子生物科学的发展,人们将会越来越多地通过分子育种技术实现快速、定向、高效选育多抗辣椒新品种。常规育种方法与分子育种方法相结合是今后的发展方向,但分子育种理论和方法还需深入研究才能达到应用程度。
4.1 分离克隆更多抗病基因并阐明其抗病分子机理分离抗病基因的目的有3个:一是可以用于分子标记辅助选择,用基因标记进行选择更精准,不会受到群体来源的影响;二是可以通过基因工程技术将抗病基因导入到感病但其他经济性状优良的自交系中;三是可以阐明抗病分子机理。
4.2 构建更精细的高密度分子标记遗传图谱辣椒的抗病性虽然通过人工接种进行鉴定和选择,但人工接种鉴定会受接种条件和发病条件的限制,还会受到主观判断认知程度的影响。当待鉴定的材料很多时,需要用到很多人工接种鉴定设施(如温室等),而分子标记鉴定可以不受时间和空间的限制,也不需要人工接种设施。但分子标记辅助选择的效率和准确率与分子标记和目标性状基因的距离有关。分子标记与目标性状基因的距离越近,分子标记辅助选择的准确率越高,受群体来源的影响也越小。只有不断提高标记密度,才能提高辅助选择的效率和准确性。
4.3 加强辣椒抗病分子生物学机理研究虽然抗病分子机理研究已经取得很多成果,但还不够深入。目前只是对生物逆境诱导前后的基因表达差异进行了研究,获得一些上调和下调表达基因,并通过正向和反向遗传学方法对部分基因进行功能鉴定。然而,由于辣椒抗病涉及到的基因很多,只是对部分基因进行功能鉴定是不够的,还无法准确掌握其调控网络,只有将生物逆境诱导前有差异表达的所有基因的功能都进行鉴定和分析,才能准确地确定其调控网络。一种病害可能由多个基因调控,一个调控基因可以调控哪些结构基因,而一个结构基因可以被哪些转录因子调控等,均需要深入研究。如果只是分离得到其中一种抗病基因,将该基因导入非抗病品种后作用不一定大,只有将抗病分子机理彻底掌握后才能从本质上解决问题。因此,今后仍需继续深入开展抗病性分子机理研究,同时还需深入了解病原菌与植物抗病性之间的关系。
4.4 加强基因编辑技术应用研究Kuroiwa等[168]采用迭代基因编辑策略(在多个位点进行编辑沉默)扩大番茄中eIF4E1的遗传多样性,并产生对多种马铃薯Y病毒分离物的抗性,有些突变体甚至达到高抗水平。在辣椒上也可以采用这种策略,一是要找到负调控基因,二是要编辑沉默多个位点,从中选择最有效的突变体。
4.5 加强辣椒转基因技术研究随着转基因产品安全性研究的不断深入,未来转基因产品可能会逐渐被人们接受。我国目前已经在多个省市试点,拟逐渐放松经证实是安全的转基因植物品种的管控力度,将来可能会有更多转基因品种用于生产。然而,目前辣椒的转基因技术还不完善,受基因型和偶然因素的影响很大,偶尔可以获得少量转基因植株,但不容易重复。在遗传转化过程中,辣椒叶丛易得,但新梢非常难,需要进一步深入研究,否则难以取得应用性成果。
| [1] |
邹学校, 胡博文, 熊程, 戴雄泽, 刘峰, 欧立军, 杨博智, 刘周斌, 索欢, 徐昊, 朱凡, 远方. 中国辣椒育种60年回顾与展望[J]. 园艺学报, 2022, 49(10): 2099-2118. DOI:10.16420/j.issn.0513-353x.2022-0677 ZOU X X, HU B W, XIONG C, DAI X Z, LIU F, OU L J, YANG B Z, LIU Z B, SUO H, XU H, ZHU F, YUAN F. Review and prospects of pepper breeding for the past 60 years in China[J]. Acta Horticulturae Sinica, 2022, 49(10): 2099-2118. DOI:10.16420/j.issn.0513-353x.2022-0677 |
| [2] |
李涛. 专题: 辣椒遗传育种与栽培生理[J]. 广东农业科学, 2023, 50(11): 9. LI T. Special topic: Genetic breeding and cultivation physiology of Capsicum[J]. Guangdong Agricultural Sciences, 2023, 50(11): 9. |
| [3] |
王立浩, 马艳青, 张宝玺. 我国辣椒品种市场需求与育种趋势[J]. 中国蔬菜, 2019(8): 1-4. WANG L H, MA Y Q, ZHANG B X. Market demand and breeding trend of pepper varieties in China[J]. China Vegetables, 2019(8): 1-4. |
| [4] |
李颖, 王恒明, 徐小万, 徐晓美, 王得元, 李乃坚, 余小林. 华南地区辣椒品种选育及育种技术研究进展[J]. 广东农业科学, 2020, 47(11): 60-69. DOI:10.16768/j.issn.1004-874X.2020.11.007 LI Y, WANG H M, XU X W, XU X M, WANG D Y, LI N J, YU X L. Breeding of pepper cultivars in South China and research progress in pepper breeding technology[J]. Guangdong Agricultural Sciences, 2020, 47(11): 60-69. DOI:10.16768/j.issn.1004-874X.2020.11.007 |
| [5] |
徐晓美, 李颖, 孙启迪, 徐小万, 衡周, 李涛, 王恒明. 辣椒种质材料疫病抗性鉴评及遗传多样性分析[J]. 广东农业科学, 2022, 49(10): 19-28. DOI: 10.16768/j.issn.1004-874X.2022.10.003. XU X M, LI Y, SUN Q D, XU X W, HENG Z, WANG H M. Resistance evaluation and genetic diversity analysis of Phytophthora disease in pepper germplasm materials[J]. Guangdong Agricultural Sciences, 2022, 49(10): 19-28. DOI: 10.16768/j.issn.1004-874X.2022.10.003. |
| [6] |
韩梅梅, 段青青, 谭延肖, 张绍丽, 李腾飞, 李华, 张超, 常培培, 王静静, 张自坤. 辣椒主要病害抗病育种研究进展[J]. 中国农学通报, 2023, 39(14): 27-32. DOI:10.11924/j.issn.1000-6850.casb2022-0438 HAN M M, DUAN Q Q, TAN Y X, ZHANG S L, LI T F, LI H, ZHANG C, CHANG P P, WANG J J, ZHANG Z K. Breeding Capsium with main disease resistance: Research progress[J]. Chinese Agricultural Science Bulletin, 2023, 39(14): 27-32. DOI:10.11924/j.issn.1000-6850.casb2022-0438 |
| [7] |
雷建军, 朱张生, 陈长明, 曹必好, 陈国菊, 郑婕, 吴昊, 肖艳辉, 蒋园园, 原远, 廖毅, 宋佳丽. 辣椒分子育种研究进展[J]. 西南大学学报: 自然科学版, 2023, 45(7): 1-20. DOI:10.13718/j.cnki.xdzk.2023.07.001 LEI J J, ZHU Z S, CHEN C M, CAO B H, CHEN G J, ZHENG J, WU H, XIAO Y H, JIANG Y Y, YUAN Y, LIAO Y, SONG J L. Progress on molecular breeding of pepper[J]. Journal of Southwest University: Natural Science Edition, 2023, 45(7): 1-20. DOI:10.13718/j.cnki.xdzk.2023.07.001 |
| [8] |
雷刚, 周坤华, 陈学军, 黄月琴, 袁欣捷, 李歌歌, 谢媛媛, 方荣. 辣椒疫病研究进展[J]. 江西农业学报, 2023, 35(6): 39-48. DOI:10.19386/j.cnki.jxnyxb.2023.06.006 LEI G, ZHOU K H, CHEN X J, HUANG Y Q, YUAN X J, LI G G, XIE Y Y, FANG R. Research progress in pepper Phytophthora blight[J]. Acta Agriculturae Jiangxi, 2023, 35(6): 39-48. DOI:10.19386/j.cnki.jxnyxb.2023.06.006 |
| [9] |
CARANTA C, PALLOIX A. Both common and specific genetic factors are involved in polygenic resistance of pepper to several potyviruses[J]. Theoretical and Applied Genetics, 1996, 92(1): 15-20. DOI:10.1007/BF00222946 |
| [10] |
HWANG J, LI J, LIU W Y, AN SJ, CHO H, HER N H, YEAM I, KIM D, KANG B C. Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper[J]. Molecules and Cells, 2009, 27: 329-336. DOI:10.1007/s10059-009-0042-y |
| [11] |
MURPHY J F, BLAUTH J R, LIVINGSTONE K D, LACKNEY V K, JAHN M K. Genetic mapping of the pvr1 locus in Capsicum spp. and evidence that distinct potyvirus resistance loci control responses that differ at the whole plant and cellular levels[J]. Molecular Plant-Microbe Interactions, 1998, 11(10): 943-951. DOI:10.1094/MPMI.1998.11.10.943 |
| [12] |
CARANTA C, THABUIS A, PALLOIX A. Development of a CAPS marker for the Pvr4 locus: A tool for pyramiding potyvirus resistance genes in pepper[J]. Genome, 1999, 42(6): 1111-1116. DOI:10.1139/gen-42-6-1111 |
| [13] |
ARNEDO-ANDRÉS M S, GIL-ORTEGA R, LUIS-ARTEAGA M, HORMAZA J I. Development of RAPD and SCAR markers linked to the Pvr4 locus for resistance to PVY in pepper (Capsicum annuum L.)[J]. Theoretical and Applied Genetics, 2002, 105(6/7): 1067-1074. DOI:10.1007/s00122-002-1058-2 |
| [14] |
TAMISIER L, SZADKOWSKI M, NEMOUCHI G, LEFEBVRE V, SZADKOWSKI E, DUBOSCQ R, SANTONI S, SARAH G, SAUVAGE C, PALLOIX A, MOURY B. Genome-wide association mapping of QTLs implied in potato virus Y population sizes in pepper: Evidence for widespread resistance QTL pyramiding[J]. Molecualr Plant Pathology, 2020, 21(1): 3-16. DOI:10.1111/mpp.12874 |
| [15] |
RUBIO M, CARANTA C, PALLOIX A. Functional markers for selection of potyvirus resistance alleles at the pvr2-eIF4E locus in pepper using tetra-primer ARMS-PCR[J]. Genome, 2008, 51(9): 767-771. DOI:10.1139/G08-056 |
| [16] |
DEVRAN Z, KAHVECI E, O ZKAYNAK E, STUDHOLME D J, TOR M. Development of molecular markers tightly linked to Pvr4 gene in pepper using next-generation sequencing[J]. Molecular Breeding, 2015, 35(4). DOI:10.1007/s11032-015-0294-5 |
| [17] |
HOLDSWORTH W L, MAZOUREK M. Development of user-friendly markers for the pvr1 and Bs3 disease resistance genes in pepper[J]. Molecular Breeding, 2015, 35(1). DOI:10.1007/s11032-015-0260-2 |
| [18] |
YEAM I, KANG B C, LINDEMAN W, FRANTZ J D, FABER N, JAHN M M. Allele-specific CAPS markers based on point mutations in resistance alleles at the pvr1 locus encoding eIF4E in Capsicum[J]. Theoretical and Applied Genetics, 2005, 112: 178-186. DOI:10.1007/s00122-005-0120-2 |
| [19] |
CARANTA C, LEFEBVRE V, PALLOIX A. Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci[J]. Molecular Plant-Microbe Interactions, 1997, 10(7): 872-878. DOI:10.1094/MPMI.1997.10.7.872 |
| [20] |
PARRELLA G, RUFFEL S, MORETTI A, MOREL C, PALLOIX A, CARANTA C. Recessive resistance genes against potyviruses are localized in colinear genomic regions of the tomato (Lycopersicon spp.) and pepper (Capsicum spp.) genomes[J]. Theoretical and Applied Genetics, 2002, 105(6/7): 855-861. DOI:10.1007/s00122-002-1005-2 |
| [21] |
VENKATESH J, AN J, KANG W H, JAHN M, KANG B C. Fine mapping of the dominant potyvirus resistance gene Pvr7 reveals a relationship with Pvr4 in Capsicum annuum[J]. Phytopathology, 2018, 108: 142-148. DOI:10.1094/PHYTO-07-17-0231-R |
| [22] |
LOAIZA-FIGUEROA F. Genetic studies on the infection-responses to tobacco mosaic and tomato spotted wilt viruses in interspecific crosses of pepper (Capsicum)[D]. New York: Cornell University, 1994.
|
| [23] |
MOURY B, PFLIEGER S, BLATTES A, LEFEBVRE V, PALLOIX A. A CAPS marker to assist selection of tomato spotted wilt virus (TSWV) resistance in pepper[J]. Genome, 2000, 43: 137-142. DOI:10.1139/g99-098 |
| [24] |
JAHN M, PARAN I, HOFFMANN K, RADWANSKI E R, LIVINGSTONE K D, GRUBE R C, AFTERGOOT E, LAPIDOT M, MOYER J. Genetic mapping of the Tsw locus for resistance to the tospovirus tomato spotted wilt virus in Capsicum spp. and its relationship to the gene for resistance to the same pathogen in tomato[J]. Molecular Plant-Microbe Interactions, 2000, 13(6): 673-82. DOI:10.1094/MPMI.2000.13.6.673 |
| [25] |
BOUKEMA I W. Allelism of genes controlling resistance to TMV in Capsicum L.[J]. Euphytica, 1980, 29: 433-439. DOI:10.1007/BF00025143 |
| [26] |
LEFEBVR E V, PALLOI X A, CARANTA C, POCHARD E. Construction of an intraspecific integrated linkage map of pepper using molecular markers and doubled-haploid progenies[J]. Genome, 1995, 38(1): 112-121. DOI:10.1139/g95-014 |
| [27] |
SUGITA T, YAMAGUCHI K, SUGIMURA Y, NAGATA R, YUJI K, KINOSHITA T. Development of SCAR markers linked to L3 gene in Capsicum[J]. Breeding Science, 2004, 54: 111-115. DOI:10.1270/jsbbs.54.111 |
| [28] |
YANG H B, LIU W Y, KANG W H, JAHN M, KANG B C. Development of SNP markers linked to the L locus in Capsicum spp. by a comparative genetic analysis[J]. Molecular Breeding, 2009, 24: 433-446. DOI:10.1007/s11032-009-9304-9 |
| [29] |
王述彬, 吴小丽, 刘金兵, 潘宝贵. 辣椒抗黄瓜花叶病毒(CMV) 基因的ISSR标记[J]. 分子植物育种, 2009, 7(3): 569-572. WANG S B, WU X L, LIU J B, PAN B G. The ISSR markers linked to CMV resistant gene in hot pepper[J]. Molecular Plant Breeding, 2009, 7(3): 569-572. |
| [30] |
杨学玲. 辣椒抗CMV基因同源序列克隆与分子标记研究[D]. 南京: 南京农业大学, 2009. YANG X L. Study on cloning of homologous sequence of resistance gene to CMV and its molecular marker in pepper[D]. Nanjing: Nanjing Agricultural Univeirsity, 2009. |
| [31] |
赵娟. 辣椒分子连锁图谱的构建及抗黄瓜花叶病毒QTL定位[D]. 呼和浩特: 内蒙古农业大学, 2009. ZHAO J. Construction of molecular linkage map and QTL analysis of cucumber mosaic virus resistance in pepper[D]. Hohhot: Inner Mongolia Agricultural University, 2009. |
| [32] |
KANG W H, HOANG N H, YANG H B, KWON J K, JO S H, SEO J K, KIM K H, CHOI D, KANG B C. Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.)[J]. Theoretical and Applied Genetics, 2010, 120: 1587-1596. DOI:10.1007/s00122-010-1278-9 |
| [33] |
KIM H, YOON J B, LEE J. Development of fluidigm SNP type genotyping assays for marker-assisted breeding of chili pepper (Capsicum annuum L.)[J]. Horticultural Science Technology, 2017, 35: 465-479. |
| [34] |
CHOI S, LEE J H, KANG W H, KIM J, HUY H N, PARK S W, SON E H, KWON J K, KANG B C. Identification of cucumber mosaic resistance 2 (cmr2) that confers resistance to a new cucumber mosaic virus isolate P1 (CMV-P1) in pepper (Capsicum spp.)[J]. Frontiers in Plant Science, 2018, 9: 1106. DOI:10.3389/fpls.2018.01106 |
| [35] |
BEN CHAIM A, GRUBE R, LAPIDOT M, JAHN M, PARAN I. Identification of quantitative trait loci associated with resistance to cucumber mosaic virus in Capsicum annuum[J]. Theoretical and Applied Genetics, 2001, 102: 1213-1220. DOI:10.1007/s001220100581 |
| [36] |
CARANTA C, PFLIEGER S, LEFEBVRE V, DAUBÈZE A M, THABUIS A, PALLOIX A. QTLs involved in the restriction of cucumber mosaic virus (CMV) long-distance movement in pepper[J]. Theoretical and Applied Genetics, 2002, 104(4): 586-591. DOI:10.1007/s001220100753 |
| [37] |
LEE H R, YOU H J, LEE Y G, KIM J, KANG H J, HARN C H B C. Development of a novel codominant molecular marker for chili veinal mottle virus resistance in Capsicum annuum L.[J]. Euphytica, 2013, 193: 197-205. DOI:10.1007/s10681-013-0897-z |
| [38] |
LEE J H, AN J T, SIDDIQUE M I, HAN K, CHOI S, KWON J K, KANG B C. Identification and molecular genetic mapping of Chili veinal mottle virus (ChiVMV) resistance genes in pepper (Capsicum annuum)[J]. Molecular Breeding, 2017, 37(10). DOI:10.1007/s11032-017-0717-6 |
| [39] |
THAKUR H, JINDAL S K, SHARMA A, DHALIWAL M S. Molecular mapping of dominant gene responsible for leaf curl virus resistance in chilli pepper (Capsicum annuum L.)[J]. 3 Biotech, 2020, 10(4). DOI:10.1007/s13205-020-02168-7 |
| [40] |
DWIVEDI N, MISHRA M, SHARMA S S, SINGH R K. Genetic analysis and QTLs identification for resistance to the Begomovirus causing pepper leaf curl virus (PepLCV) disease[J]. Journal of Plant Biochemistry and Biotechnology, 2024, 33(1): 34-44. DOI:10.1007/s13562-023-00855-z |
| [41] |
MIMURA Y, KAGEYAMA T, MINAMIYAMA Y, HIRAI M. QTL analysis for resistance to Ralstonia solanacearum in Capsicum accession 'LS2341'[J]. Journal of Japan Society of Horticultural Science, 2009, 78: 307-313. DOI:10.2503/jjshs1.78.307 |
| [42] |
THAKUR P P, MATHEW D, NAZEEM P A, ABIDA P S, INDIRA P, GIRIJA D, SHYLAJA M R, VALSALA A. Identification of allele specific AFLP markers linked with bacterial wilt ﹝ Ralstonia solanacearum (Smith) Yabuuchi et al. ﹞resistance in hot peppers (Capsicum annuum L.)[J]. Physiological and Molecular Plant Pathology, 2014, 87: 19-24. DOI:10.1016/j.pmpp.2014.05.001 |
| [43] |
KANG Y J, AHN Y K, KIM K T, JUN T H. Resequencing of Capsicum annuum parental lines (YCM334 and Taean) for the genetic analysis of bacterial wilt resistance[J]. BMC Plant Biology, 2016, 16. DOI:10.1186/s12870-016-0931-0 |
| [44] |
LEE S, CHAKMA N, JOUNG S, LEE J M, LEE J. QTL mapping for resistance to bacterial wilt caused by two isolates of Ralstonia solanacearum in chili pepper (Capsicum annuum L.)[J]. Plants-Basel, 2022, 11(12). DOI:10.3390/plants11121551 |
| [45] |
CHAE S Y, LEE K, DO J W, HONG S C, LEE K H, CHO M C, YANG E Y, YOON J B. QTL mapping of resistance to bacterial wilt in pepper plants (Capsicum annuum) using genotyping-by-sequencing (GBS)[J]. Horticulturae, 2022, 8(2). DOI:10.3390/horticulturae8020115 |
| [46] |
马海宾. 辣椒抗疫病的生化和分子标记研究[D]. 儋州: 华南热带农业大学, 2002. MA H B. Studies on biochemical marker of pepper blight resistance and RAPD Marker linked to Phytophthora capsici gene in Capsicum [D]. Danzhou: South China Tropical Agricultural University, 2002. |
| [47] |
易图永. 辣椒抗疫病相关基因的分析及QTL定位[D]. 长沙: 湖南农业大学, 2003. YI T Y. Analysis and QTL mapping of the relative resistant gene to Phtophthora blight in pepper(Capsicum annuum)[D]. Changsha: Hunan Agricultrual University, 2003. |
| [48] |
安静. 辣椒分子连锁图谱的构建及抗疫病QTL定位[D]. 北京: 中国农业科学院, 2006. AN J. Construction of molecular linkage map and QTL analysis of Phytophthora capsici resistance in pepper[D]. Beijing: Chinese Academy of Agricultural Sciences, 2006. |
| [49] |
李儒剑. 辣椒抗疫病分子标记的开发及抗病相关基因的功能鉴定[D]. 杨凌: 西北农林科技大学, 2018. LI R J. Development of molecular markers and functional identification of disease-resistant genes in pepper[D]. Yangling: Northwest Agricultural and Forestry University, 2018. |
| [50] |
LI Y F, ZHANG S C, YANG X M, WANG C P, HUANG Q Z, HUANG R Z. Generation of a high-density genetic map of pepper (Capsicum annuum L.) by SLAF-seq and QTL analysis of Phytophthora capsici resistance[J]. Horticulturae, 2021, 7(5). DOI:10.3390/horticulturae7050092 |
| [51] |
KUMAR M, KAMBHAM M R, REDDY D C L, SRIRAM S, SINGH T H. Identification of molecular marker linked to resistance gene loci against Indian isolate of Phytophthora capsici L. causing root rot in chilli (Capsicum annuum L.)[J]. Australasian Plant Pathology, 2021, 51(2): 211-220. DOI:10.1007/s13313-021-00837-6 |
| [52] |
LOZADA D N, NUNEZ G, LUJAN P, DURA S, COON D, BARCHENGER D W, SANOGO S, BOSLAND P W. Genomic regions and candidate genes linked with Phytophthora capsici root rot resistance in chile pepper (Capsicum annuum L.)[J]. BMC Plant Biology, 2021, 21(1). DOI:10.1186/s12870-021-03387-7 |
| [53] |
BONGIORNO G, DI N A, CIANCALEONI S, MARCONI G, CASSIBBA V, ALBERTINI E. Development and application of a cleaved amplified polymorphic sequence marker (Phyto) linked to the Pc5.1 locus conferring resistance to Phytophthora capsici in pepper (Capsicum annuum L.)[J]. Plants-Basel, 2023, 12(15). DOI:10.3390/plants12152757 |
| [54] |
ZHANG Z H, CAO Y C, WANG Y F, YU H L, WU H M, LIU J, AN D L, ZHU Y S, FENG X G, ZHANG B X, WANG L H. Development and validation of KASP markers for resistance to Phytophthora capsici in Capsicum annuum L.[J]. Molecular Breeding, 2023, 43(3). DOI:10.1007/s11032-023-01367-3 |
| [55] |
RO N, HAILE M, HUR O, GEUM B, RHEE J, HWANG A, KIM B, LEE J, HAHN B S, LEE J, KANG B C. Genome-wide association study of resistance to Phytophthora capsici in the pepper (Capsicum spp.) collection[J]. Frontiers in Plant Science, 2022, 13. DOI:10.3389/fpls.2022.902464 |
| [56] |
MOHAMMADBAGHERI L, NASR-ESFAHANI M, AL-SADI A M, KHANKAHDANI H H, GHADIRZADEH E. Screening for resistance and genetic population structure associated with Phytophthora capsici-pepper root and crown rot[J]. Physiological and Molecular Plant Patholog, 2022, 122. DOI:10.1016/j.pmpp.2022.101924 |
| [57] |
BUKHARI T, RANA R M, UL HASSAN M, NAZ F, SAJJAD M. Genetic diversity and marker trait association for Phytophthora resistance in chilli[J]. Molecular Biology Reports, 2022, 49(6): 5717-5728. DOI:10.1007/s11033-022-07635-3 |
| [58] |
LIU W Y, KANG J H, JEONG H S, CHOI H J, YANG H B, KIM K T, CHOI D, CHOI G J, JAHN M, KANG B C. Combined use of bulked segregant analysis and microarrays reveals SNP markers pinpointing a major QTL for resistance to Phytophthora capsici in pepper[J]. Theoretical and Applied Genetetics, 2014, 127: 2503-2513. DOI:10.1007/s00122-014-2394-8 |
| [59] |
ATES A Ç, YILMAZ N. molecular marker assisted selection for Phytophthora capsici Leon. resistance lines in pepper (Capsicum annum L.)[J]. Acta Scientiarum Polonorum-Hortorum cultus, 2020, 19(3): 179-188. DOI:10.24326/asphc.2020.3.16 |
| [60] |
THABUIS A, PALLOIX A, PFLIEGER S, DAUBÈZE A M, CARANTA C, LEFEBVRE V. Comparative mapping of Phytophthora resistance loci in pepper germplasm: Evidence for conserved resistance loci across Solanaceae and for a large genetic diversity[J]. Theoretical and Applied Genetics, 2003, 106(8): 1473-1485. DOI:10.1007/s00122-003-1206-3 |
| [61] |
THABUIS A, LEFEBVRE V, BERNARD G, DAUBÈZE A M, PHALY T, POCHARD E, PALLOIX A. Phenotypic and molecular evaluation of a recurrent selection program for a polygenic resistance to Phytophthora capsici in pepper[J]. Theoretical and Applied Genetics, 2004, 109(2): 342-351. DOI:10.1007/s00122-004-1633-9 |
| [62] |
LEE J, HONG J, DO J W. Identification of QTLs for resistance to anthracnose to two Colletotrichum species in pepper[J]. Journal of Crop Sciences Biotechnology, 2010, 13: 227-233. DOI:10.1007/s12892-010-0081-0 |
| [63] |
LEE J, DO J W, YOON J B. Development of STS markers linked to the major QTLs for resistance to the pepper anthracnose caused by Colletotrichum acutatum and C. capsici[J]. Horticultural Environment Biotechnology, 2011, 52(596): 601. DOI:10.1007/s13580-011-0178-5 |
| [64] |
NANDA C, PRATHIBHA V H, RAO A M, RAMESH S, HITTALMANI S, PAI S. Tagging SSR markers associated with genomic regions controlling anthracnose resistance in chilli[J]. Indian Journal of Horticulture, 2016, 73(3): 350-355. DOI:10.5958/0974-0112.2016.00076.1 |
| [65] |
SUWOR P, THUMMABENJAPONE P, SANITCHON J, KUMAR S, TECHAWONGSTIEN S. Phenotypic and genotypic responses of chili (Capsicum annuum L.) progressive lines with different resistant genes against anthracnose pathogen(Colletotrichum spp.)[J]. European Journal of Plant Pathology, 2015, 143: 725-736. DOI:10.1007/s10658-015-0723-7 |
| [66] |
MISHRA R, ROUT E, MOHANTY J N, JOSHI R K. Sequence-tagged site-based diagnostic markers linked to a novel anthracnose resistance gene RCt1 in chili pepper (Capsicum annuum L.)[J]. 3 Biotech, 2019, 9(1). DOI:10.1007/s13205-018-1552-0 |
| [67] |
ZHAO Y, LIU Y, ZHANG Z, CAO Y, YU H, MA W, ZHANG B, WANG R, GAO J, WANG L. Fine mapping of the major anthracnose resistance QTL AnRGO5 in Capsicum chinense 'PBC932'[J]. BMC Plant Biology, 2020, 20: 189. DOI:10.1186/s12870-019-2115-1 |
| [68] |
SUWOR P, SANITCHON J, THUMMABENJAPONE P, KUMAR S, TECHAWONGSTIEN S. Inheritance analysis of anthracnose resistance and marker-assisted selection in introgression populations of chili (Capsicum annuum L.)[J]. Scientia Horticulturae, 2017, 22: 20-26. DOI:10.1016/j.scienta.2017.03.032 |
| [69] |
MAHASUK P, STRUSS D, MONGKOLPORN O. QTLs for resistance to anthracnose identified in two Capsicum sources[J]. Molcular Breeding, 2016, 36: 10. DOI:10.1007/s11032-016-0435-5 |
| [70] |
CHEN Y, ZENG Q, MAN Y L, LIU S Z, OUYANG C, LI C G, WU X Y, ZHANG D Y, LIU Y, TAN X Q. Simple sequence repeat markers reflect the biological phenotype differentiation and genetic diversity of Colletotrichum gloeosporioides strains from Capsicum annuum L. in China[J]. Journal of Phytopathology, 2021, 169(11/12): 701-709. DOI:10.1111/jph.13041 |
| [71] |
VOORRIPS R E, FINKERS R, SANJAYA L, GROENWOLD R. QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annum and C. chinense[J]. Theoretical and Applied Genetics, 2004, 109: 1275-1282. DOI:10.1007/s00122-004-1738-1 |
| [72] |
SUN C Y, MAO S L, ZHANG Z H, PALLOIX A, WANG L H, ZHANG B X. Resistances to anthracnose (Colletotrichum acutatum) of Capsicum mature green and ripe fruit are controlled by a major dominant cluster of QTLs on chromosome P5[J]. Scientia Horticulturae, 2015, 181(2): 81-88. |
| [73] |
RO N, HAILE M, HUR O, KO H C, YI J Y, WOO H J, CHOI Y M, RHEE J, LEE Y J, KIM D A, DO J W, KIM G W, KWON J K, KANG B C. Genome-wide association study of resistance to anthracnose in pepper (Capsicum chinense) germplasm[J]. BMC Plant Biology, 2023, 23(1). DOI:10.1186/s12870-023-04388-4 |
| [74] |
TAI T H, DAHLBECK D, CLARK E T, GAJIWALA P, PASION R, WHALEN M C, STALL R E, STASKAWICZ B J. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(24): 14153-1458. DOI:10.1073/pnas.96.24.14153 |
| [75] |
TRUONG H T H, KIM K T, KIM S, CHO M C, KIM H R, WOO J G. Development of gene-based markers for the Bs2 bacterial spot resistance gene for marker-assisted selection in pepper (Capsicum spp.)[J]. Horticulture, Environment, and Biotechnology, 2011, 52: 65-73. DOI:10.1007/s13580-011-0142-4 |
| [76] |
PIERRE M, NOEL L, LAHAYE T, BALLVORA A, VEUSKENS J, GANAL M, BONAS U. High-resolution genetic mapping of the pepper resistance locus Bs3 governing recognition of the Xanthomonas campestris pv vesicatora AvrBs3 protein[J]. Theoretical and Applied Genetics, 2000, 101: 255-263. DOI:10.1007/s001220051477 |
| [77] |
SHARMA A, MINSAVAGE G, GILL U S, HUTTON S F, JONES J B. Identification and mapping of bs8, a novel locus conferring resistance to bacterial spot caused by Xanthomonas gardneri[J]. Phytopathology, 2022, 112(8): 1640-1650. DOI:10.1094/PHYTO-08-21-0339-R |
| [78] |
SHARMA A, LI J, WENTE R, MINSAVAGE G V, GILL U S, ORTEGA A, VALLEJOS C E, HART J P, STASKAWICZ B J, MAZOUREK M R, STALL R E, JONES J B, HUTTON S F. Mapping of the bs5 and bs6 non-race-specific recessive resistances against bacterial spot of pepper[J]. Frontiers in Plant Science, 2023, 14. DOI:10.3389/fpls.2023.1061803 |
| [79] |
王立浩, 张宝玺, CARANTA C, 毛胜利, PALLOIX A. 利用分子标记对辣椒抗马铃薯Y病毒的3个QTLs进行选择[J]. 园艺学报, 2008, 35(1): 53-58. DOI:10.16420/j.issn.0513-353x.2008.01.011 WANG L H, ZHANG B X, CARANTA C, MAO S L, PALLOIX A. Molecular markers assisted selection for three QTLs resistant to PVY in pepper (Capsicum annuum L.)[J]. Acta Horticultrae Sinica, 2008, 35(1): 53-58. DOI:10.16420/j.issn.0513-353x.2008.01.011 |
| [80] |
MOODLEY V, NAIDOO R, GUBBA A, MAFONGOYA P L. Development of potato virus Y (PVY) resistant pepper (Capsicum annuum L.) lines using marker-assisted selection (MAS)[J]. Physiological and Molecular Plant Pathology, 2019, 105(96/101). DOI:10.1016/j.pmpp.2018.12.002 |
| [81] |
于海龙, 靳远, 周黛媛, 张正海, 曹亚从, 吴华茂, 冯锡刚, 张宝玺, 王秀芝, 崔聪聪, 王立浩. 内蒙古地区辣椒种质资源抗病性鉴定与评价[J]. 中国蔬菜, 2023(11): 69-79. DOI:10.19928/j.cnki.1000-6346.2023.2041 YU H L, JIN Y, ZHOU D Y, ZHANG Z H, CAO Y C, WU H M, FENG X G, ZHANG B X, WANG X Z, CUI C C, WANG L H. Identification and evaluation of disease resistance in pepper germplasms from Inner Mongolia Autonomous Region[J]. China Vegetable, 2023(11): 69-79. DOI:10.19928/j.cnki.1000-6346.2023.2041 |
| [82] |
李子雄, 潘兵青, 宋莹莹, 马世杰, 张娣, 陈婕, 沈火林, 孙亮. 辣椒种质资源果实品质与抗病性综合评价[J]. 中国蔬菜, 2023(8): 46-58. DOI:10.19928/j.cnki.1000-6346.2023.2033 LI Z X, PAN B Q, SONG Y Y, MA S J, ZHANG D, CHEN J, SHEN H L, SUN L. Comprehensive evaluation on fruit quality and disease resistance of pepper germplasm resources[J]. China Vegetable, 2023(8): 46-58. DOI:10.19928/j.cnki.1000-6346.2023.2033 |
| [83] |
陈灵芝, 张茹, 王兰兰, 高彦萍. 与辣椒抗TMV L3基因连锁的分子标记的筛选及种质资源抗TMV鉴定[J]. 西北农业学报, 2023, 32(4): 585-592. DOI:10.7606/j.issn.1004-1389.2023.04.011 CHEN L Z, ZHANG R, WANG L L, GAO Y P. Application of tobamvirus reistance gene L3-linked markers in pepper (Capsicum spp.) resource[J]. Acta Agriculturae Boreali-occidengtalis Sinica, 2023, 32(4): 585-592. DOI:10.7606/j.issn.1004-1389.2023.04.011 |
| [84] |
陈建分, 曹振木, 秦于玲, 申龙斌, 刘维侠, 朱丹, 吴怡婷, 刘子记, 王旭. 辣椒种质资源PMMoV抗性基因检测与抗性鉴定[J/OL]. 热带作物学报, 2023, https://kns.cnki.net/kcms/detail/46.1019.s.20230316.1409.004.html. CHEN J F, CAO Z M, QIN Y L, SHEN L B, LIU W X, ZHU D, WU Y T, LIU Z J, WANG X. Resistance gene detection and resistance identification of PMMoV in pepper germplasm[J/OL]. Chinese Journal of Tropical Crops, 2023, https://kns.cnki.net/kcms/detail/46.1019.s.20230316.1409.004.html. |
| [85] |
FIDAN H, YILDIZ K, SARIKAYA P. Molecular detection of resistance-breaking strain Cucumber mosaic virus (rbCMV) (Cucumovirus; Bromoviridae) on resistant commercial pepper cultivars in Turkey[J]. Journal of Phytopathology, 2023, 171(6): 234-241. DOI:10.1111/jph.13175 |
| [86] |
OZKAYNAK E, DEVRAN Z, KAHVECI E, DOGANLAR S, BASKÖYLÜ B, DOGAN F, ISLEYEN M, YÜKSEL A, YÜKSEL M. Pyramiding multiple genes for resistance to PVY, TSWV and PMMoV in pepper using molecular markers[J]. European Journal of Horticultural Science, 2014, 79(4): 233-239. DOI:10.2307/24126862 |
| [87] |
POLAT E, OZALP R. Molecular marker assisted selection for resistance to tomato spotted wilt virus (TSWV) in pepper breeding[J]. Journal of Biotechnology, 2015, 185: 114. DOI:10.1016/j.jbiotec.2014.07.387 |
| [88] |
SOOD T, SOOD S, SOOD V K, BADIYAL A, ANURADHA, KAPOOR S. Assessment and validation of resistance to bacterial wilt (Ralstonia solanacearum) through field and molecular studies in bell pepper[J]. Journal of Plant Pathology, 2023, 105(3): 849-857. DOI:10.1007/s42161-023-01378-1 |
| [89] |
郭爽, 黄贞, 常绍东, 刘玉平, 曹翠文. 利用分子标记鉴定辣椒抗疫病材料[J]. 中国农学通报, 2012, 28(13): 163-166. GUO S, HUANG Z, CHANG S D, LIU Y P, CAO C W. Identification of resistance to phytophthora blight in hot pepper using molecular marker[J]. Chinese Agricultural Science Bulletin, 2012, 28(13): 163-166. |
| [90] |
ATES A Ç, YILMAZ N. Molecular marker assisted selection for Phytophthora capsici Leon. resistance lines in pepper (Capsicum annuum L.)[J]. Acta Scientiarum Polonorum-Hortorum Cultus, 2020, 19(3): 179-188. DOI:10.24326/asphc.2020.3.16 |
| [91] |
RABUMA T, GUPTA O P, CHHOKAR V. Phytophthora capsici infection and analysis of genetic diversity among identified resistance accessions using SSR markers[J]. Physiological and Molecular Plant Pathology, 2020, 112(1). DOI:10.1016/j.pmpp.2020.101539 |
| [92] |
KIM H, YOON J B, LEE J. Development of fluidigm SNP type genotyping assays for marker-assisted breeding of chili pepper (Capsicum annuum L.)[J]. Horticultural Science Technology, 2017, 35: 465-479. |
| [93] |
DING L N, LI Y T, WU Y Z, LI T, GENG R, CAO J, ZHANG W, TAN X L. Plant disease resistance-related signaling pathways: Recent progress and future prospects[J]. International Journal of Molecular Sciences, 2022, 23(24). DOI:10.3390/ijms232416200 |
| [94] |
DANG F F, WANG Y N, SHE J J, LEI Y F, LIU Z Q, EULGEM T, LAI Y, LIN J, YU L, LEI D, GUAN D Y, LI X, YUAN Q, HE S L. Overexpression of CaWRKY27, a subgroup Ⅱe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection[J]. Physiologia Plantarum, 2014, 150(3): 397-411. DOI:10.1111/ppl.12093 |
| [95] |
MOU S L, LIU Z Q, GAO F, YANG S, SU M X, SHEN L, WU Y, HE S L. CaHDZ27, a homeodomain-leucine zipper I protein, positively regulates the resistance to Ralstonia solanacearum infection in pepper[J]. Molecular Plant-microbe Interactions, 2017, 30(12): 960-973. DOI:10.1094/MPMI-06-17-0130-R |
| [96] |
CHENG W, XIAO Z L, CAI H Y, WANG C Q, HU Y, XIAO Y P, ZHENG Y X, SHEN L, YANG S, LIU Z Q, MOU S L, QIU A L, GUAN D Y, HE S L. A novel leucine-rich repeat protein, CaLRR51, acts as a positive regulator in the response of pepper to Ralstonia solanacearum infection[J]. Molecular Plant Pathology, 2017, 18(8): 1089-1100. DOI:10.1111/mpp.12462 |
| [97] |
HUANG J F, SHEN L, YANG S, GUANG D Y, HE S L. CaASR1 promotes salicylic acid-but represses jasmonic acid-dependent signaling to enhance the resistance of Capsicum annuum to bacterial wilt by modulating CabZIP63[J]. Journal of Experimental Botany, 2020, 71(20): 6538-6554. DOI:10.1093/jxb/eraa350 |
| [98] |
YANG S, SHI Y Y, ZOU L Y, HUANG J F, SHEN L, WANG Y Z, GUAN D Y, HE S L. Pepper CaMLO6 negatively regulates Ralstonia solanacearum resistance and positively regulates high temperature and high humidity responses[J]. Plant Cell Physiology, 2020, 61(7): 1223-1238. DOI:10.1093/pcp/pcaa052 |
| [99] |
YANG S, ZHANG Y W, CAI W W, LIU C L, HU J, SHEN L, HUANG X Y, GUAN D Y, HE S L. CaWRKY28 Cys249 is required for interaction with CaWRKY40 in the regulation of pepper immunity to Ralstonia solanacearum[J]. Molecular Plant-microbe Interactions, 2021, 34(7): 733-745. DOI:10.1094/MPMI-12-20-0361-R |
| [100] |
SHEN L, YANG S Y, FENG F, GUAN D Y, HE S L. CaCBL1 acts as a positive regulator in pepper response to Ralstonia solanacearum[J]. Molecular Plant-microbe Interactions, 2020, 33(7): 945-957. DOI:10.1094/MPMI-08-19-0241-R |
| [101] |
SHI L P, LI X, WENG Y H, CAI H Y, LIU K S, XIE B X, ANSAR H, GUAN D Y, HE S L, LIU Z Q. The CaPti1-CaERF3 module positively regulates resistance of Capsicum annuum to bacterial wilt disease by coupling enhanced immunity and dehydration tolerance[J]. Plant Journal, 2022, 111(1): 250-268. DOI:10.1111/tpj.15790 |
| [102] |
HUSSAIN A, KAISHENG L, NOMAN A, ASHRAF M F, ALBAQAMI M, KHAN M I, LIU Z Q, HE S L. N-Methyltransferase CaASHH3 acts as a positive regulator of immunity against bacterial pathogens in pepper[J]. International Journal of Molecular Sciences, 2022, 23(12). DOI:10.3390/ijms23126492 |
| [103] |
YANG S, CAI W W, SHEN L, CAO J S, LIU C L, HU J, GUAN D Y, HE S L. A CaCDPK29-CaWRKY27b module promotes CaWRKY40-mediated thermotolerance and immunity to Ralstonia solanacearum in pepper[J]. New Phytologist, 2022, 233(4): 1843-1863. DOI:10.1111/nph.17891 |
| [104] |
LU Q L, HUANG Y, WANG H, WAN M Y, LYU J G, CHENG X G, CHEN Y H, CAI W W, YANG S, SHEN L, GUAN D Y, HE S L. CabZIP23 integrates in CabZIP63-CaWRKY40 cascade and turns CabZIP63 on mounting pepper immunity against Ralstonia solanacearum via physical interaction[J]. International Journal of Molecular Science, 2022, 23: 2656. DOI:10.3390/ijms23052656 |
| [105] |
ZHANG Y Q, GUO S Y, ZHANG F, GAN P F, LI M, WANG C, LI H K, GAO G, WANG X J, KANG Z S, ZHANG X M. CaREM1.4 interacts with CaRIN4 to regulate Ralstonia solanacearum tolerance by triggering cell death in pepper[J]. Horticulture Research, 2023, 10(5): uhad053. DOI:10.1093/hr/uhad053 |
| [106] |
YANG S, CAI W W, WU R J, HUANG Y, LU Q L, HUANG X Y, ZHANG Y P, WU Q, CHENG X G, WAN M Y, LYU J A, LIU Q, ZHENG X, MOU S L, GUAN D Y, HE S L. Differential CaKAN3-CaHSF8 associations underlie distinct immune and heat responses under high temperature and high humidity conditions[J]. Nature Communications, 2023, 14(1): 4477. DOI:10.1038/s41467-023-40251-8 |
| [107] |
ZHANG Y P, CAI W W, WANG A W, HUANG X Y, ZHENG X, LIU Q, CHENG X G, WAN M Y, LYU J A, GUAN D Y, YANG S, HE S L. MADS-box protein AGL8 interacts with chromatin-remodelling component SWC4 to activate thermotolerance and environment-dependent immunity in pepper[J]. Journal of Experimental Botany, 2023, 74(12): 3667-3683. DOI:10.1093/jxb/erad092 |
| [108] |
AMBARWATI E, ARWIYANTO T, WIDADA J, ALAM T, ANDIKA I, TARYONO. The genes associated with jasmonic acid and salicylic acid are induced in tropical chili pepper against Ralstonia solanacearum by applying Arbuscular Mycorrhizal fungi[J]. Horticulturae, 2022, 8(10). DOI:10.3390/horticulturae8100876 |
| [109] |
WANG J E, LIU K K, LI D W, ZHANG Y L, ZHAO Q, HE Y M, GONG Z H. A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophtora capsici infection as well as abiotic stress tolerance[J]. International Journal of Molecular Sciences, 2013, 14(2): 3158-3177. DOI:10.3390/ijms14023158 |
| [110] |
LIU Z Q, SHI L P, YANG S, LIN Y Q, WENG Y H, LI X, HUSSAIN A, NOMAN A, HE S L. Functional and promoter analysis of Chi IV3, a chitinase of pepper plant, in response to Phytophthora capsici infection[J]. International Journal of Molecular Sciences, 2017, 18(8): 1661. DOI:10.3390/ijms18081661 |
| [111] |
ALI M, MUHAMMAD I, HAQ S U, ALAM M, KHATTAK A M, AKHTAR K, ULLAH H, KHAN A, LU G, GONG Z H. The CaChiVI2 gene of Capsicum annuum L. confers resistance against heat stress and infection of Phytophthora capsici[J]. Frontier in Plant Science, 2020, 11. DOI:10.3389/fpls.2020.00219 |
| [112] |
JIN J H, ZHANG H X, TAN J Y, YAN M J, LI D W, KHAN A, GONG Z H. A new ethylene-responsive factor CaPTI1 gene of pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici[J]. Frontiers in Plant Science, 2016(6): 1217. DOI:10.3382/fpis.2015.01217 |
| [113] |
JIN J H, ZHANG H X, ALI M, WEI A M, LUO D X, GONG Z H. The CaAP2/ERF064 regulates dual functions in pepper: Plant cell death and resistance to Phytophthora capsici[J]. Genes, 2019, 10(7): 541. DOI:10.3390/genes10070541 |
| [114] |
ZHANG H X, FENG X H, ALI M, JIN J H, WEI A M, KHATTAK A M, GONG Z H. Identification of pepper CaSBP08 gene in defense response against Phytophthora capsici infection[J]. Frontiers in Plant Science, 2020(11): 183. DOI:10.3389/fpls.2020.00183 |
| [115] |
ZHANG H X, FENG X H, JIN J H, KHAN A, GUO W L, DU X H, GONG Z H. CaSBP11 participates in the defense response of pepper to Phytophthora capsici through regulating the expression of defense-related genes[J]. International Journal of Molecular Sciences, 2020, 21: 9065. DOI:10.3390/ijms21239065 |
| [116] |
ZHANG H X, ALI M, FENG X H, JIN J H, HUANG L J, KHAN A, LYU J G, GAO S Y, LUO D X, GONG Z H. A novel transcription factor CaSBP12 gene negatively regulates the defense response against Phytophthora capsici in pepper (Capsicum annuum L.)[J]. International Journal of Molecular Sciences, 2019, 20(48). DOI:10.3390/ijms20010048 |
| [117] |
ZHANG Y L, JIA Q L, LI D W, WANG J E, YIN Y X, GONG Z H. Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici Leonian[J]. International Journal of Molecular Sciences, 2013, 14(5): 8985-9004. DOI:10.3390/ijms14058985 |
| [118] |
KHAN A, LI R J, SUN J T, MA F, ZHANG H X, JIN J H, ALI M UL HAQ S, WANG J E, GONG Z H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses[J]. Scientific Reports, 2018, 8(1): 5500. DOI:10.1038/s41598-018-23761-0 |
| [119] |
MA X, GAI W X, QIAO Y M, ALI M, WEI A M, LUO D X, LI Q H, GONG Z H. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.)[J]. BMC Genomics, 2019, 20(1): 775. DOI:10.1186/s12864-019-6125-z |
| [120] |
CHENG W, JIANG Y, PENG J T, GUO J W, LIN M L, JIN C T, HUANG J F, TANG W Q, GUAN D Y, HE S L. The transcriptional reprograming and functional identification of WRKY family members in pepper's response to Phytophthora capsici infection[J]. BMC Plant Biology, 2020, 20: 1. DOI:10.1186/s12870-020-02464-7 |
| [121] |
KANG W H, KIM S, LEE H A, CHOI D, YEOM S I. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper[J]. Scientific Reports, 2016, 6: 33332. DOI:10.1038/srep33332 |
| [122] |
DU J S, HANG L F HAO H Q, YANG H T, ALI S, BADAWY R S E, XU X Y, TAN H Q, SU L H, LI H X, ZOU K X, LI Y, SUN B, LIN L J, LAI Y S. The dissection of R genes and locus Pc5.1 in Phytophthora capsici infection provides a novel view of disease resistance in peppers[J]. BMC Genomics, 2021, 372. DOI:10.1186/s12864-021-07705-z |
| [123] |
NABOR-ROMERO O, ZAVALETA-MEJIA E, OCHOA-MARTINEZ D L, SILVA-VALENZUELA M, VEGA-ARREGUIN J, SANCHEZFLORES A, ROJAS-MARTINEZ R I. Transcriptional alterations induced by Nacobbus aberrans in interaction with chili pepper CM-334 and Phytophthora capsici[J]. Physiological and Molecular Plant Pathology, 2023, 123. DOI:10.1016/j.pmpp.2022.101942 |
| [124] |
ZHOU L Y, YANG S Z, CHEN C L, LI M, DU Q J, WANG J Q, YIN Y X, XIAO H J. CaCP15 gene negatively regulates salt and osmotic stress responses in Capsicum annuum L.[J]. Genes, 2023, 14(7): 1409. DOI:10.3390/genes14071409 |
| [125] |
BABA V Y, POWELL A F, IVAMOTO-SUZUKI S T, PEREIRA L F P, VANZELA A L L, GIACOMIN R M, STRICKLER S R, MUELLER L A, RODRIGUES R, GONCALVES L S A. Capsidiol-related genes are highly expressed in response to Colletotrichum scovillei during Capsicum annuum fruit development stages[J]. Scientic Reports, 2020, 10(1): 12048. DOI:10.1038/s41598-020-68949-5 |
| [126] |
LEE S C, HWANG I S, CHOI H W, HWANG B K. Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance[J]. Plant Physiology, 2008, 148(2): 1004-1020. DOI:10.1104/pp.108.123836 |
| [127] |
SON S, KIM S, LEE K S, OH J, CHOI I, DO J W, YOON J B, HAN J, PARK S R. The Capsicum baccatum-specific truncated NLR protein CbCN enhances the innate immunity against Colletotrichum acutatum[J]. International Journal of Molecular Sciences, 2021, 22(14): 7672. DOI:10.3390/ijms22147672 |
| [128] |
LI Y, MA X, XIAO L D, YU Y N, YAN H L, GONG Z H. CaWRKY50 acts as a negative regulator in response to Colletotrichum scovillei infection in pepper[J]. Plants, 2023, 12: 1962. DOI:10.3390/plants12101962 |
| [129] |
HAN Y J, KANG H Y, KIM Y S, KIM J I. Functional divergence of two closely related carboxylesterases in pepper (Capsicum annuum L.)[J]. Plant Biotechnology Reports, 2023, 17(4): 499-507. DOI:10.1007/s11816-023-00849-2 |
| [130] |
SRIDEEPTHI R, KRISHNA M S R, SUNEETHA P, KRISHNA R S, KARTHIKEYAN S. Genome-wide identification, characterization and expression analysis of non-RD receptor like kinase gene family under Colletotrichum truncatum stress conditions in hot pepper[J]. Genetica, 2020, 148: 283-296. DOI:10.1007/s10709-020-00104-4 |
| [131] |
SON S, KIM S, LEE K S, OH J, CHOI I, DO J W, YOON J B, HAN J, CHOI D, PARK S R. Identification of the Capsicum baccatum NLR protein CbAR9 conferring disease resistance to anthracnose[J]. International Journal of Molecular Sciences, 2021, 22(22): 12612. DOI:10.3390/ijms222212612 |
| [132] |
MISHRA R, MOHANTY J N, MAHANTY B, JOSHI R K. A single transcript CRISPR/Cas9 mediated mutagenesis of CaERF28 confers anthracnose resistance in chilli pepper (Capsicum annuum L.)[J]. Planta, 2021, 254: 5-17. DOI:10.1007/s00425-021-03660-x |
| [133] |
SHAFIQUE S, SHAFIQUE S, AHMAD A. Biochemical and molecular screening of varieties of chili plants that are resistant against Fusarium wilt infection[J]. European Journal of Microbiology and Immunology, 2018, 8: 12-19. DOI:10.1556/1886.2017.00031 |
| [134] |
HONG J K, HWANG B K. Functional characterization of PR-1 protein, β-1, 3-glucanase and chitinase genes during defense response to biotic and abiotic stresses in Capsicum annuum[J]. The Plant Pathology Journal, 2005, 21: 195-206. DOI:10.5423/PPJ.2005.21.3.195 |
| [135] |
KIM D S, KIM N H, HWANG B K. The Capsicum annuum class Ⅳ chitinase ChitⅣ interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses[J]. Journal of Experimental Botany, 2015, 66: 1987-1999. DOI:10.1093/jxb/erv001 |
| [136] |
KOEDA S, ONOUCHI M, MORI N, POHAN N S, NAGANO A J, KESUMAWATI E. A recessive gene pepy-1 encoding Pelota confers resistance to begomovirus isolates of PepYLCIV and PepYLCAV in Capsicum annuum[J]. Theoretical and Applied Genetics, 2021, 134(9): 2947-2964. DOI:10.1007/s00122-021-03870-7 |
| [137] |
OH S K, LEE S, YU S H, CHOI D. Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens[J]. Planta, 2005, 222(5): 876-887. DOI:10.1007/s00425-005-0030-1 |
| [138] |
ZHAO L H, ZHANG L Z, HU Z H, LI B W, ZHENG X, QIU R S, CHEN Y, LI J, DONG J H, ZHANG Z K. Tomato zonate spot virus induced hypersensitive resistance via an auxin-related pathway in pepper[J]. Gene, 2022, 823: 146320. DOI:10.1016/j.gene.2022.146320 |
| [139] |
LUO Y, QIN C, QIU H R, ZHANG X W, TANG X Q, LUO X R, LUO Y, YANG H, CHEN X C. Novel microRNAs associated with the immune response to cucumber mosaic virus in hot pepper (Capsicum annuum L.)[J]. Physiological and Molecular Plant Pathology, 2023, 124. DOI:10.1016/j.pmpp.2023.101963 |
| [140] |
HAN K L, ZHENG H Y, YAN D K, ZHOU H J, JIA Z X, ZHAI Y S, WU J, LU Y W, WU G W, RAO S F, CHEN J P, PENG J J, QI R D, YAN F. Pepper mild mottle virus coat protein interacts with pepper chloroplast outer envelope membrane protein OMP24 to inhibit antiviral immunity in plants[J]. Horticulture Research, 2023, 10(5). DOI:10.1093/hr/uhad046 |
| [141] |
WANG J, ZENG X, TIAN D S, YANG X B, WANG L L, YIN Z C. The pepper Bs4C proteins are localized to the endoplasmic reticulum (ER) membrane and confer disease resistance to bacterial blight in transgenic rice[J]. Molecular Plant Pathology, 2018, 19(8): 2025-2035. DOI:10.1111/mpp.12684 |
| [142] |
SENDIN L N, ORCE I G, GOMEZ R L, ENRIQUE R, BOURNONVILLE C F G, NOGUERA A S, VOJNOV A A, MARANO M R, CASTAGNARO A P, FILIPPONE M P. Inducible expression of Bs2 R gene from Capsicum chacoense in sweet orange (Citrus sinensis L. Osbeck) confers enhanced resistance to citrus canker disease[J]. Plant Molecular Biology, 2017, 93(6): 607-621. DOI:10.1007/s11103-017-0586-8 |
| [143] |
SZABO Z, BALOGH M, DOMONKOS A, CSANYI M, KALO P, KISS G B. The bs5 allele of the susceptibility gene Bs5 of pepper (Capsicum annuum L.) encoding a natural deletion variant of a CYSTM protein conditions resistance to bacterial spot disease caused by Xanthomonas species[J]. Theoretical and Applied Genetics, 2023, 136(3): 64. DOI:10.1007/s00122-023-04340-y |
| [144] |
RAFFEINER M, USTUN S, GUERRA T, SPINTI D, FITZNER M, SONNEWALD S, BALDERMANN S, BORNKE F. The Xanthomonas type-Ⅲ effector XopS stabilizes CaWRKY40a to regulate defense responses and stomatal immunity in pepper (Capsicum annuum)[J]. Plant Cell, 2022, 34(5): 1684-1708. DOI:10.1093/plcell/koac032 |
| [145] |
DJAMI-TCHATCHOU A T, MATSAUNYANE L B T, KALU C M, NTUSHELO K. Gene expression and evidence of coregulation of the production of some metabolites of chilli pepper inoculated with Pectobacterium carotovorum ssp. Carotovorum[J]. Functional Plant Biology, 2019, 46(12): 1114-1122. DOI:10.1071/FP18244 |
| [146] |
GER M J, LOUH G Y, LIN Y H, FENG T Y, HUANG H E. Ectopically expressed sweet pepper ferredoxin PFLP enhances disease resistance to Pectobacterium carotovorum subsp. carotovorum affected by harpin and protease-mediated hypersensitive response in Arabidopsis[J]. Molecular Plant Pathology, 2014, 15(9): 892-906. DOI:10.1111/mpp.12150 |
| [147] |
CHEN Z L, GU H Y, LI Y, SU Y L, WU P, JIANG Z C, MING X T, TIAN J H, PAN N S, QU L J. Safety assessment for genetically modified sweet pepper and tomato[J]. Toxicology, 2003, 188: 297-307. DOI:10.1016/S0300-483X(03)00111-2 |
| [148] |
商鸿生, 王旭, 徐秉良. 转CP基因线辣椒对CMV的抗病性组分研究[J]. 西北农业学报, 2001, 10(1): 29-32. SHANG H S, WANG X, XU B L. The components of coat protein-mediated resistance to CMV in transgenic chili pepper[J]. Acta Agriculturae Boreali Occidentalis Sinica, 2001, 10(1): 29-32. |
| [149] |
徐秉良, 商鸿生, 王旭. 双抗转CP基因线辣椒对非克隆病毒的抗病性[J]. 兰州大学学报, 2002, 38(4): 95-100. XU B L, SHANG H S, WANG X. Protection of transgenic chili pepper expressing cmv and TMV coat proteins gene against unrelated CMV strains and viruses[J]. Journal of Lanzhou University (Natural Sciences), 2002, 38(4): 95-100. |
| [150] |
徐秉良, 商鸿生, 王旭. 转CP基因线辣椒对CMV和CMV-RNA的抗病性比较[J]. 植物病理学报, 2002, 32(2): 132-137. DOI:10.13926/j.cnki.apps.2002.02.006 XU B L, SHANG H S, WANG X. Comparison of resistance to CMV particle and to CMV-RNA in transgenic chili pepper expressing cmv and tmv coat proteins[J]. Acta Phytopathologica Sinica, 2002, 32(2): 132-137. DOI:10.13926/j.cnki.apps.2002.02.006 |
| [151] |
杨国顺. 转JERFs基因提高辣椒抗病性的研究[D]. 长沙: 湖南农业大学, 2003. YANG G S. Studies on disease resistance of transgenic pepper with jasmonate and ethylene responsive element bingding factor genes[D]. Changsha: Hunan Agricultural University, 2003. |
| [152] |
高玉尧, 陈长明, 陈国菊, 曹必好, 雷建军. Cry2Aa2和PamPAP双价表达载体的构建及其对辣椒的遗传转化[J]. 园艺学报, 2012, 39(7): 1285-1292. DOI:10.16420/j.issn.0513-353x.2012.07.013 GAO Y Y, CHEN C M, CHEN G J, CAO B H, LEI J J. Construction of binary expression vector containing Cry2Aa2 and PamPAP and its transfer to pepper[J]. Acta Horticulturae Sinica, 2012, 39(7): 1285-1292. DOI:10.16420/j.issn.0513-353x.2012.07.013 |
| [153] |
黄真池. 诱导超敏反应增强辣椒广谱抗病性的转基因研究[D]. 长沙: 湖南农业大学, 2008. HUANG C. Transgene studies on enhancing pepper broad-spectrum disease resistance by inducing hypersensitive response[D]. Changsha: Hunan Agricultural University, 2008. |
| [154] |
BAGGA S, LUCERO Y, APODACA K, RAJAPAKSE W, LUJAN P, ORTEGA J L, SENGUPTA-GOPALAN C. Chile (Capsicum annuum) plants transformed with the RB gene from Solanum bulbocastanum are resistant to Phytophthora capsici[J]. PLOS One, 2019, 14(10). DOI:10.1371/journal.pone.0223213 |
| [155] |
周钟信, 粟密兰, 陈德芬, 宋兰英, 杨静惠. 辣椒诱导再生及黄瓜花叶病毒外壳基因转化研究初报[J]. 华北农学报, 1991, 6(14): 69-72. ZHOU Z X, LI M L, CHEN D F, SONG L Y, YANG J H. Preliminary study on regeneration in duction and gene transformation of CMVcp in pepper[J]. Acta Agriculaturae Boreali Sinica, 1991, 6(14): 69-72. |
| [156] |
张宗江, 周钟信, 刘艳军, 江倩云, 尤明, 刘国民, 米景九. 黄瓜花叶病毒壳蛋白基因转化辣椒及其在转基因株后代的表达[J]. 华北农学报, 1994, 9(3): 67-71. ZHANG Z J, LIU Y J, JIANG Q Y, YOU M, LIU G M, MI J J. CMVcp gene transforamtion and expression in pepper[J]. Acta Agriculaturae Boreali Sinica, 1994, 9(3): 67-71. |
| [157] |
李华平, 胡晋生, 王敏, 范怀忠. 黄瓜花叶病毒衣壳蛋白基因转化辣椒研究[J]. 病毒学报, 2000, 16(3): 276-278. DOI:10.13242/j.cnki.bingduxuebao.001258 LI H P, HU J S, WANG M, FAN H Z. Studies on transgenic pepper plants transferred with the coat protein gene of cucumber mosaic virus[J]. Chinese Journal of Virology, 2000, 16(3): 276-278. DOI:10.13242/j.cnki.bingduxuebao.001258 |
| [158] |
XU B L, SHENG H S, WANG X. Comparison of resistance to CMV particle and to CMV-RNA in transgenic chili pepper expressing CMV and TMV coat proteins[J]. Acta Phytopatholgoica Sinica, 2002, 32(2): 132-137. |
| [159] |
毕玉平, 单蕾, 王兴军, 徐平丽, 周钟信, 米景九. 双抗TMV+CMV辣椒转基因工程植株的再生及抗病毒鉴定[J]. 华北农学报, 1999, 14(3): 103-108. BI Y P, SHAN L, WANG X J, XU L P, ZHOU Z X, MI J J. Regeneration of transgenic pepper plants resistant to TMV and CMV and its resistance assay[J]. Acta Agriculturae Boreali Sinica, 1999, 14(3): 103-108. |
| [160] |
郭亚华, 徐香玲, 邓立平, 张军民, 张欣. Ri质粒介导TMV和CMV外壳蛋白基因转化甜椒研究[J]. 北方园艺, 2000(4): 17-18. GUO Y H, XI X L, DENG L P, ZHANG J M, ZHANG X. Study on transformation of TMV and CMV coat protein genes mediated by Ri plasmid in sweet pepper[J]. Northern Horticulture, 2000(4): 17-18. |
| [161] |
陈国菊, 石丽, 雷建军, 曹必好, 曾国平. 中国商陆抗病毒蛋白基因的克隆及其转化辣椒[J]. 园艺学报, 2008, 35(6): 827-832. DOI:10.16420/j.issn.0513-353x.2008.06.013 CHEN G J, SHI L, LEI J J, CAO B H, ZENG G P. Cloning of pokeweed antiviral protein gene from Phytolacca acinosa and its transfer to pepper (Capsicum annuum L.)[J]. Acta Horticulturae Sinica, 2008, 35(6): 827-832. DOI:10.16420/j.issn.0513-353x.2008.06.013 |
| [162] |
MURPHY J F, KYLE M M. Isolation and viral-infection of Capsicum leaf protoplasts[J]. Plant Cell Reports, 1994, 13(7): 397-400. DOI:10.1007/BF00234146 |
| [163] |
董春枝, 姜春哓, 冯兰香. 甜(辣) 椒导入CMV卫星RNA互补DNA的植株再生[J]. 园艺学报, 1992, 19(2): 184-186. DONG C Z, JIANG C X, FENG L X. Transgenic pepper plants (Capsicum annuum L.) containing CMV Sat-RNA cDNA[J]. Acta Horticulturae Sinica, 1992, 19(2): 184-186. |
| [164] |
KIM Y H. Improvementin plant disease resistance and anti-fungal protein gene[J]. Proceedings Vienna Austria, 1995, 7: 145-155. |
| [165] |
张银东, 唐跃东, 曾宪松. 抗菌肽基因转化辣椒的研究[J]. 华南热带农业大学学报, 2000, 6(1): 1-4. ZHANG Y D, TANG Y D, ZENG X S. Study on transformation of antimicrobial peptide gene in pepper[J]. Journal of South China University of Tropical Agriculture, 2000, 6(1): 1-4. |
| [166] |
李乃坚, 余小林, 李颖, 黄自然, 张银东. 双价抗菌肽基因转化辣椒[J]. 热带作物学报, 2000, 21(4): 45-51. LI N J, YU X L, LI Y, HUANG Z R, ZHANG Y D. Transference of double gene cecropin B and D into pepper (Capsicum annuum L.)[J]. Chinese Journal of Tropical Crops, 2000, 21(4): 45-51. |
| [167] |
李颖, 余小林, 李乃坚, 王恒明, 黄自然. 转抗菌肽基因辣椒株系的青枯病抗性鉴定及系统选育[J]. 分子植物育种, 2005, 3(2): 217-221. LI Y, YU X L, LI N J, WANG H M, HUANG Z R. Evaluation of resistance to bacterial wilt and systemic breeding in cecropin-GM Capsicum[J]. Molecular Plant Breeding, 2005, 3(2): 217-221. |
| [168] |
KUROIWA K, DANILO B, PERROT L, THENAULT C, VEILLET F, DELACOTE F, DUCHATEAU P, NOGUE, MAZIER M, GALLOIS J L. An iterative gene-editing strategy broadens eIF4E1 genetic diversity in Solanum lycopersicum and generates resistance to multiple potyvirus isolates[J]. Plant Biotechnology Journal, 2023. DOI:10.1111/pbi.14003 |
(责任编辑 张辉玲)
 2024, Vol. 51
2024, Vol. 51